Abstract
Rejuvenation refers to the transition from an adult state to a juvenile state. Trunk truncation at the base of the tree can result in tree rejuvenation. However, little is known about the association of rejuvenation with leaf biomass and flavonoid accumulation. The results of this study showed that, compared with control leaves, leaves of renewed Ginkgo biloba shoots were larger, thicker, and more lobed and had higher fresh/dry weights and chlorophyll contents. The leaf biomass per hectare of rejuvenated trees was twofold higher than that of the untruncated controls. Moreover, we observed a marked increase in the accumulation of flavonol glycosides via metabolomic analysis and detected upregulated expression of genes involved in flavonoid biosynthesis, including CHS, FLS, F3’H, DFR, and LAR. Overexpression of GbCHS in ginkgo calli confirmed that GbCHS plays an important role in flavonoid biosynthesis. Interestingly, the contents of gibberellins significantly increased in the rejuvenated leaves. Moreover, exogenous gibberellin treatment significantly increased GbCHS expression and flavonoid contents. Our findings show that truncation can stimulate tree rejuvenation by altering hormone levels, representing an effective and feasible approach for enhancing the biomass and flavonoid content of G. biloba leaves.
Introduction
Tree growth and development involve two distinct phases, the juvenile phase and mature phase, which differ in terms of tree phenotype, structure, and physiology [1–4]. The programmed transition from juvenility to maturity is mainly driven by age and environmental conditions. However, since trees have pools of meristematic cells in their organs [2, 3], different developmental stages (juvenile and adult) can co-occur in the same tree [3–7]. In general, the base of the tree trunk shows more juvenile characteristics than the top does [3, 4, 7]. Rejuvenation, also known as juvenile–mature phase reversal, can reverse the chronological phase transition by inducing the formation of adventitious buds and release of dormant buds [5, 6, 8]. In fact, this is a frequent occurrence in horticultural plant species, whereas complete or partial rejuvenation, often through cutting-based propagation, pruning, and coppicing, contributes to the reinvigoration of old or nonvigorous trees [9]. Rejuvenation accompanied by organ regeneration usually causes changes in organ morphology, including increased leaf size and stronger rooting [2, 10].
Rejuvenation is regulated by endogenous hormones such as auxin, cytokinins, gibberellins (GAs), and abscisic acid (ABA) [6, 10]. The ratio of two phytohormones, auxin and cytokinin, determines the developmental fate of regenerating tissue [6, 11], whereas GAs play important roles in maintaining the juvenile characteristics of plants [6, 10–13]. In maize (Zea mays), reduced levels of endogenous GAs delay the transition from the juvenile to adult vegetative state [14, 15]. Similarly, in Arabidopsis and rice, GA loss-of-function mutants exhibited prolonged juvenile phases [16, 17]. Certain age-regulated miRNAs, such as microRNA156 (miR156) and microRNA172 (miR172), are involved in the regulation of shoot regenerative capacity [18–20]. Overexpression of miR156, which reduces the expression of its target SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes, delays the juvenile-to-adult transition in both transgenic Arabidopsis thaliana and Populus × canadensis [7, 21]. Interestingly, crosstalk between GA and miR156 mediated the floral transition of Arabidopsis thaliana [22]. In horticultural trees, miR156 expression was shown to decrease as trees age and is correlated with the developmental phase transition [23]. In contrast, miRNA172 was shown to downregulate the APETALA2-like gene to promote the juvenile-to-adult phase change in plants [18]. In addition, as mobile signals, sugars promote the juvenile-to-adult transition by reducing miR156 levels [24, 25]. Several sugar signaling molecules promote the expression of miR156 and thereby delay vegetative phase change [24–26].
Flavonoids are a major class of secondary metabolic products that play vital roles in regulating cell physiology, signaling, and the interaction between plants and the external environment [27]. Flavonoid biosynthesis pathways have been extensively characterized in model plant species. Many structural and regulatory genes involved in the flavonoid biosynthesis pathway have been identified, such as those encoding chalcone synthase (CHS), flavonol synthase (FLS), MYBs, basic helix–loop–helix (bHLH) proteins, and WD40 proteins [28, 29]. Importantly, a direct link between different developmental phases and secondary metabolism has been demonstrated. miR156-targeted SPLs could affect the age-dependent accumulation of secondary metabolites [30, 31]. In addition, anthocyanin accumulation occurs in accordance with age-regulated miRNAs; for example, increased miR156 activity promotes the accumulation of anthocyanins, whereas reduced miR156 activity results in high levels of flavonols [32]. Although these studies indicated an association of plant age with flavonoid accumulation, the effect of rejuvenation on flavonoid biosynthesis in trees is unknown. In addition, how rejuvenation reverses age-dependent traits in primitive gymnosperms is unclear.
Ginkgo biloba is an important tree species with substantial medicinal and ecological value [33, 34]. The leaves of G. biloba contain an abundance of active compounds, such as flavonoids, terpene lactones, polysaccharides, and phenols, among which more than 40 types of flavonol glycosides have been identified [35]. Flavonoids are the main bioactive ingredients in G. biloba extract (GbE) [36, 37]. GbE is largely harvested from leaves and has been developed as a standardized formulation phytomedicine named EGb761, which is used in the treatment of cardiovascular, neurodegenerative, and aging-related diseases, among others [37–39]. GbE is also sold as a dietary supplement and natural health product in many countries [39, 40]. GbE has become one of the most popular and well-researched herbal preparations worldwide [36, 37], resulting in ginkgo becoming an industrial crop species with high market demand. Several studies have shown that the content of flavonoids in G. biloba leaves is closely related to tree age, with higher flavonoid glycoside contents occurring in saplings (≤ 5 years) than in adult trees [41]. Consequently, the decrease in flavonoids as trees age negatively influences the medicinal quality of the leaves. However, how truncation affects tree rejuvenation and leaf flavonoid content in G. biloba is unknown.
Our main objectives were to investigate the effects of truncation on G. biloba tree rejuvenation, as well as leaf biomass and leaf flavonoid content. We applied a rejuvenation technique to 5-year-old G. biloba trees and analyzed leaf morphology and size, fresh and dry weight (DW), mesophyll cell structure, and photochemical efficiency. Using RNA sequencing (RNA-seq) and metabolomics, we characterized the gene–metabolite regulatory network of flavonoid accumulation in rejuvenated plants. By combining endogenous hormone analysis with exogenous hormone treatment validation, we revealed that the hormone changes following truncation were responsible for rejuvenation and contributed to the accumulation of biomass and flavonoids in the leaves. This truncation technique can be used to generate an optimized tree form and increase yield and quality during the commercial production of leaf products.
Results
Morphological, cytological, and physiological changes in leaves following truncation
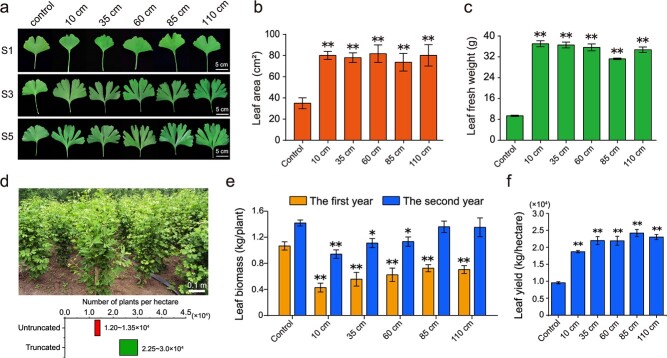
We applied stem truncation treatments to 5-year-old healthy G. biloba trees, cutting them back to trunk heights of 10, 35, 60, 85, and 110 cm (Fig. 1). After the treatments, numerous sprouts were produced from the trunks (Fig. 1b–h), which then developed into juvenile branches (Fig. 1i–m). The sprouts near the cutting position developed particularly rapidly and formed longer lateral branches and larger leaves than did those near the base of the trunk, which grew slowly and produced smaller leaves (Fig. S1).
Figure 1.
Morphological changes of G. biloba under the indicated truncation treatments. a Five-year-old G. biloba trees at S1. b–f Trees under the indicated truncation treatments at S1. The red arrows indicate sprouting. g–h Local features of the truncation position. The blue arrow indicates the truncation position on the trunk. The red arrows indicate sprouting close to the truncation position. i–m Trees at S2 after truncation to heights of 10 cm (i), 35 cm (j), 60 cm (k), 85 cm (l), and 110 cm (m).
Compared to the leaves of the untruncated controls, the leaves of trees exhibited significant variations in morphology, cytology, and physiology according to truncation height. Because the leaf lobes of G. biloba are important morphological characteristics associated with the juvenile state [42, 43], we evaluated the number of lobes on rejuvenated and control leaves. The truncated plants produced highly lobed leaves with an average of 6–7 lobes, far more than the two lobes typically produced by the controls (Fig. 2a, Fig. S2a). Similarly, the leaves of all truncated plants became thicker; for example, the leaf thickness under the 10 cm treatment was 4.3 ± 0.05 mm compared to 2.6 ± 0.05 mm for the controls, representing an increase of approximately 50% (Fig. S2b); this was greater than the increase in the other truncated plants. Moreover, the trees in all the truncation groups showed more than a twofold increase in leaf area relative to that of the controls (Fig. 2a, b). The fresh and DWs of the leaves in all the truncation groups exhibited a three- or fourfold increase compared to those of the controls (Fig. 2c, Fig. S2c). The leaf water content was at least 71.5% in the truncation groups, whereas the average water content in the controls was 65.9 ± 0.4% (Fig. S2d). Moreover, we determined the leaf yield per plant for two years after truncation (Fig. 2e). The leaf yield (0.46–0.73 kg per plant) in the first year decreased significantly in the truncation groups compared with the controls, whereas a substantial increase (0.95–1.35 kg per plant) was observed during the second year. Applying the truncation technique, we maintained a high density of 5-year-old G. biloba trees for sustainable harvesting. Due to the decrease in tree size after truncation, a higher density of 22 500–30 000 trees could be planted per hectare (Fig. 2d) compared to the 12 500–13 500 trees that can be cultivated without truncation (traditional leaf harvesting ginkgo farm). The leaf yields per hectare of the truncation groups were more than twofold that of the untruncated controls, reflecting increased leaf yield per hectare of the treated G. biloba trees (Fig. 2f). Because the truncation treatments of G. biloba trees were conducted every other year, this technique can ensure long-term commercial output for leaf harvesting ginkgo farms.
Figure 2.
Physiological changes in rejuvenated leaves. a Leaves subjected to truncation treatments at S1 (young), S3 (mature stage), and S5 (leaf-harvesting stage). Scale, 5 cm. b–c Leaf area (b) and leaf fresh weight (c) during the leaf-harvesting period following the truncation treatments. d Estimated numbers of trees that can be planted per hectare with and without truncation. e Leaf biomass (kg/plant) in the 2 years following treatment. f Leaf yield (kg/hectare) after truncation treatment. Shown are the means ± SDs (n = 30 for leaf biomass; n > 30 for other physiological measurements). * and ** indicate significant differences relative to the controls at p < 0.05 and p < 0.01, respectively, as determined by ANOVA with post hoc tests.
In September, at the leaf-harvesting stage, the chlorophyll content (SPAD value) of the leaves differed between the truncation groups, ranging from 64.91 to 70.95, and was significantly higher than that of the controls (58.31; Fig. S2e). The Fv/Fm of the leaves in the truncation groups presented slightly higher but not significantly different levels of photochemical efficiency relative to those of the controls (Fig. S2f). Furthermore, ultrastructural observations of the mesophyll cells in the leaves of the truncated trees showed an intact chloroplast structure at the leaf-harvesting stage (Fig. S2g), whereas the organelles had begun to degrade in the controls, showing loosely arranged organelles, distorted chloroplast lamellae, and an accumulation of osmiophilic granules, indicating delayed senescence in rejuvenated leaves (Fig. S2h).
Changes in the secondary metabolites of rejuvenated leaves
To evaluate the secondary metabolites of G. biloba leaves under the truncation treatments, we measured the leaf flavonoid and terpene lactone contents. Compared to that in the controls (2.01 ± 0.081 mg/g), the total content of terpene lactones in the taller truncated trees (truncated to 60, 85, or 110 cm) significantly increased by approximately 28–34% (Fig. 3a), reaching 2.57 ± 0.062 mg/g in the 65 cm treatment group, 2.66 ± 0.076 mg/g in the 85 cm treatment group, and 2.69 ± 0.094 mg/g in the 110 cm treatment group. In particular, the contents of ginkgolides A and B increased significantly in all truncation groups (Fig. 3b, c). Similarly, the total flavonoid contents were at least 14% higher in the truncation treatments, with the 85 and 110 cm treatments displaying substantial increases (Fig. 3d). Based on our evaluation of leaf yield and active constituent contents, we suggest that 85 and 110 cm truncations are optimum.
Figure 3.
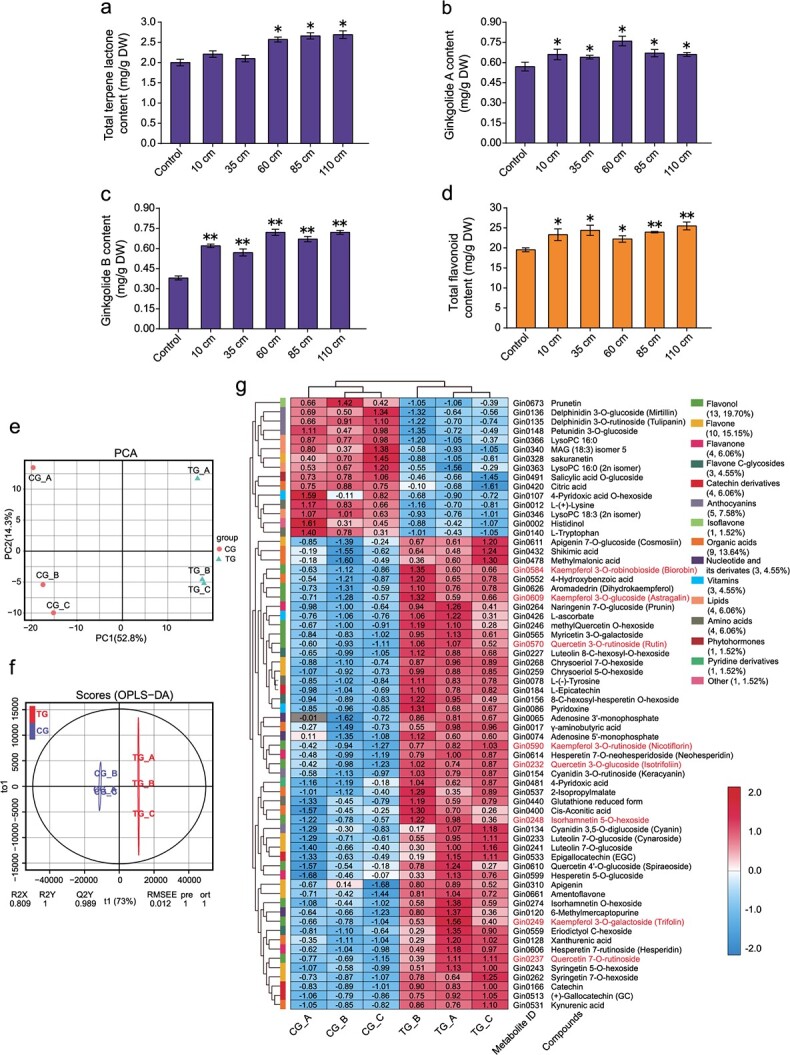
Metabolite analysis of rejuvenated leaves at the leaf-harvesting stage. a Total contents of terpene lactones following truncation treatments. DW, dry weight. b–c Contents of ginkgolide A (b) and ginkgolide B (c) after the truncation treatments. d Total content of flavonoids after the truncation treatments, as determined by a plant flavonoid test kit. * and ** indicate significant differences relative to the controls at p < 0.05 and p < 0.01, respectively, as determined by ANOVA with post hoc tests. e Principal component analysis score plot of the metabolomes of six individual samples collected from control (CG) and truncated (TG) trees. The x-axis represents PC1, and the y-axis represents PC2. f Orthogonal projections to latent structures–discriminant analysis (OPLS–DA) score plots showing the significant differences in metabolic profiles between the control and treatment groups. RMSEE, root mean square error of estimation. g Hierarchical cluster and quantitative analyses of the differentially abundant metabolites via a heatmap based on the metabolome platform. The differentially abundant metabolites are grouped together (number and percentage data are shown). The numerical values are the log10(peak area) of the metabolites in each sample. The red color indicates a high metabolite content, and the blue indicates a low metabolite content. Kaempferol, quercetin, and isorhamnetin metabolites are marked in red.
Using widely targeted metabolomics, we further analyzed leaf flavonoids in a truncation group (85 cm) and the control group. Principal component analysis (PCA) showed that the first two principal components (PC1 and PC2) explained 52.8% and 14.3% of the metabolic variance in all the samples (Fig. 3e), with most of the metabolic variance between the control and truncation plants being explained by PC1. Score plots of orthogonal projections to latent structures–discriminant analysis (OPLS–DA) further indicated that the metabolites were significantly different between the rejuvenated and control leaves (Fig. 3f). We detected a total of 692 metabolites spanning 34 categories among all samples, of which 66 significantly varied between the controls and the treatments. Of those 66 metabolites, 51 (77.3%) were upregulated and 15 (12.7%) were downregulated in the rejuvenated leaves (Fig. 3g, Table S1). These differentially abundant metabolites were grouped into 15 categories, with flavonoid-related components, including flavonols, flavones, flavanones, flavone C-glycosides, isoflavones, catechins, and anthocyanins, accounting for 60.6% (40/66) of the total. Importantly, more than 87% (35/40) of the differentially abundant flavonoid metabolites were significantly increased in the truncation group, including flavonols (quercetin glycosides, kaempferol glycosides, isorhamnetin glycosides) and flavones (apigenin, luteolin glycosides). Based on a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, these differentially abundant metabolites were enriched in the biosynthesis of secondary metabolites, flavonoid biosynthesis, and flavone and flavonol biosynthesis pathways, further demonstrating that rejuvenation promoted the accumulation of flavonoid metabolites in leaves.
We constructed a network of the correlations between the differentially abundant metabolites by calculating the Pearson correlation coefficient (r2 ≥ 0.9, p < 0.05) of the metabolite–metabolite correlations (Fig. S3). In total, 796 significant correlations were calculated, with values ranging from −0.9978 to 0.9987; 572 (71.86%) were significantly positive, and 224 (28.14%) were significantly negative (Fig. S3a). We further summarized these correlations based on the metabolite categories and found that 40 flavonoid metabolites were involved in 702 correlations (518 positive and 184 negative), which was the largest role any group played in the metabolite correlations (Fig. S3b). Moreover, 343 (300 positive and 43 negative) of the 702 correlations were between two flavonoids, indicating that the accumulation of these flavonoid components was closely related in G. biloba leaves. Furthermore, 103 correlations were between a flavonoid and organic acid, among which 86 were positive. The metabolite group showing the second highest number of correlations comprised nine organic acids involved in 154 significant correlations, including 116 positive and 38 negative correlations (Fig. S3b); 15 of those correlations (10 positive and 5 negative) were between two organic acids. Similarly, the amino acid and lipid groups showed significant correlations related to flavonoids and organic acids.
Changes in transcripts and metabolites involved in flavonoid biosynthesis in rejuvenated leaves
To investigate the molecular changes coordinating flavonoid biosynthesis, we performed RNA-seq using the same leaf samples as those used in the metabolomic analysis. In total, 26 832 and 27 192 genes were expressed in the control and rejuvenated leaves, respectively (Fig. S4a), of which 1151 and 1511 were unique to the control and rejuvenated leaves, respectively. A total of 1051 genes were differentially expressed between the treatments, comprising 254 downregulated genes and 797 upregulated genes in the rejuvenated leaves (Fig. S4b). Gene Ontology (GO) and KEGG enrichment analyses of the differentially expressed genes (DEGs) revealed that a large number of genes were significantly enriched in the phenylpropanoid biosynthesis and flavonoid biosynthesis processes (Fig. S4c, d). Most of the DEGs (21/23) related to phenylpropanoid biosynthesis were upregulated in the rejuvenation sample, including Gb_09283 (PEROXIDASE 12 [PER12]), Gb_11540 (PER21), Gb_20206 (PER42), Gb_04735 (CAFFEOYL SHIKIMATE ESTERASE [CSE]), and Gb_18171 (CAFFEOYL-COA O-METHYLTRANSFERASE [CCOAOMT]) (Fig. 4a). Intriguingly, all DEGs involved in flavonoid biosynthesis, especially Gb_19002 (CHALCONE SYNTHASE [CHS]), showed significant increases in expression in rejuvenated leaves.
Figure 4.
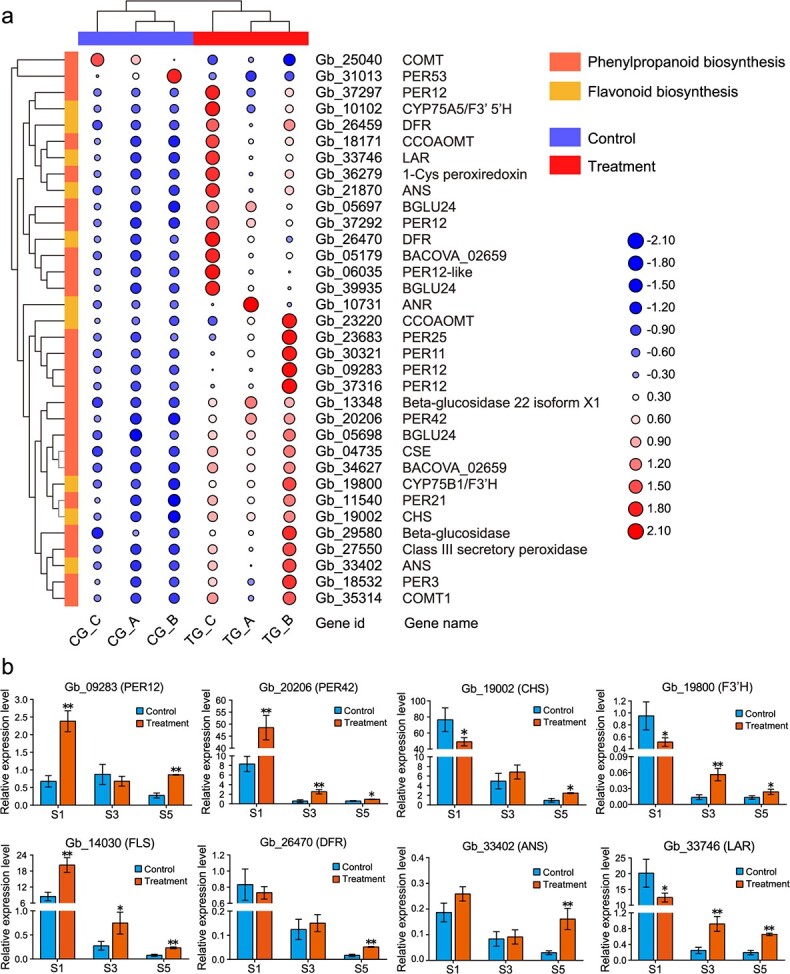
Differential expression of genes involved in secondary metabolite pathways in rejuvenated leaves. a Hierarchical clustering heatmap of differentially expressed genes related to phenylpropanoid biosynthesis and flavonoid biosynthesis. The value matrix is indicated by both the continuous color scheme and the circle size. The expression values in a row are normalized. The original numbers of fragments per kilobase of transcript per million mapped reads are displayed in the corresponding circles. b Results of a qRT–PCR analysis of the expression profiles of eight candidate genes involved in phenylpropanoid (PER12, PER42) and flavonoid biosynthesis (DFR, F3’H, ANS, LAR, FLS, and CHS) in S1, S3 and S5. The data are the means ± SDs of relative expression levels (n = 3). * and ** indicate significant differences from the controls at p < 0.05 and p < 0.01, respectively, as determined by Student’s t-test. ANS, ANTHOCYANIDIN SYNTHASE; CHS, CHALCONE SYNTHASE; DFR, DIHYDROFLAVONOL 4-REDUCTASE; F3′H, FLAVONOID 3′-HYDROXYLASE; FLS, FLAVONOL SYNTHASE; LAR, LEUCOANTHOCYANIDIN REDUCTASE; PER12, PEROXIDASE 12; PER42, PEROXIDASE 42.
Quantitative real-time PCR (qRT–PCR) was employed to examine the dynamic expression changes of eight selected DEGs associated with phenylpropanoid (PER12 and PER42) and flavonoid biosynthesis (CHS, ANTHOCYANIDIN SYNTHASE [ANS], DIHYDROFLAVONOL 4-REDUCTASE [DFR], LEUCOANTHOCYANIDIN REDUCTASE [LAR], and FLAVONOID 3′-HYDROXYLASE [F3’H/CYP75B1]) in leaves at the young (S1), mature (S3), and leaf-harvesting (S5) stages of development (Fig. 4b). The expression level of PER42 significantly increased in the rejuvenated leaves at all three stages, whereas that of PER12 significantly increased only at S1 and S5. The expression levels of CHS, F3’H, DFR, ANS, and LAR were significantly higher in the rejuvenated samples than in the control samples from S3 to S5, and FLS (Gb_14030) expression was significantly increased in the truncation sample in all three stages. In addition, three SPL genes were evaluated; the expression levels of SPL3 (Gb_23724), SPL8 (Gb_22010), and SPL12 (Gb_27198) decreased at S1 but significantly increased at S5 in the truncated trees (Fig. S6). In general, the expression of 10 of the 11 genes was generally consistent with the RNA-seq data, despite some differences in expression levels.
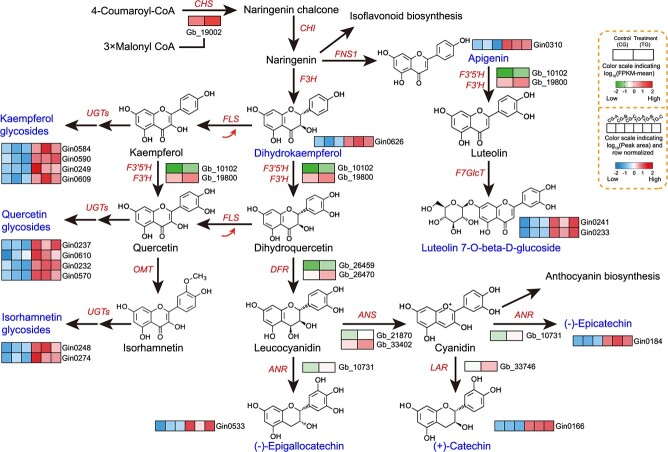
We identified the potential flavonoid biosynthetic pathway in G. biloba , integrating the changes in metabolite contents and the expression of biosynthesis-related genes (Fig. 5). Upregulation of CHS, FLS, FLAVONOID 3′5′-HYDROXYLASE (F3’5’H), F3’H, DFR, ANTHOCYANIDIN REDUCTASE (ANR), ANS, and LAR in the truncation group contributed to the increased contents of downstream apigenin and dihydrokaempferol (CHS), kaempferol and quercetin glycosides (FLS), luteolin 7-O-D-glucoside (F3′5′H and F3′H), epigallocatechin and epicatechin (DFR, ANR, and ANS), and catechin (LAR).
Figure 5.
Putative flavonoid biosynthetic pathway in G. biloba. The proposed metabolic pathways were drawn based on information in the literature and the KEGG database of metabolic pathways. The differentially abundant metabolites and differentially expressed genes between the control and truncation treatments are listed in blue and red text, respectively.
Association of transcripts and metabolites reveals that GbCHS is a hub gene in flavonoid biosynthesis
We modeled the key players in flavonoid biosynthesis by constructing a subnetwork of transcript–metabolite correlations (Fig. 6a). All differentially abundant transcripts and metabolites involved in flavonoid biosynthesis, as well as transcripts related to the phenylpropanoid pathway, were subjected to correlation analysis. A correlation network was constructed, consisting of 46 nodes and 132 edges. A total of 122 phenylpropanoid or flavonoid biosynthesis-related gene–metabolite correlations were significantly positive, and only 10 were significantly negative. CHS was found to be a hub gene that connected 28 flavonoid metabolites, of which 89.3% (25/28) were positively correlated, reflecting its importance in the flavonoid network. Moreover, F3’H, DFR, LAR, ANS, and CCOAOMT were secondary hub genes and showed mostly significant positive correlations with metabolites. Nevertheless, some metabolites, including delphinidin 3-O-rutinoside (an anthocyanin), prunetin (an isoflavone), and petunidin 3-O-glucoside (an anthocyanin), were negatively correlated with the biosynthesis-related genes CHS, ANS, DFR, and F3’H, indicating that these flavonoids are also involved in other secondary metabolic pathways.
Figure 6.
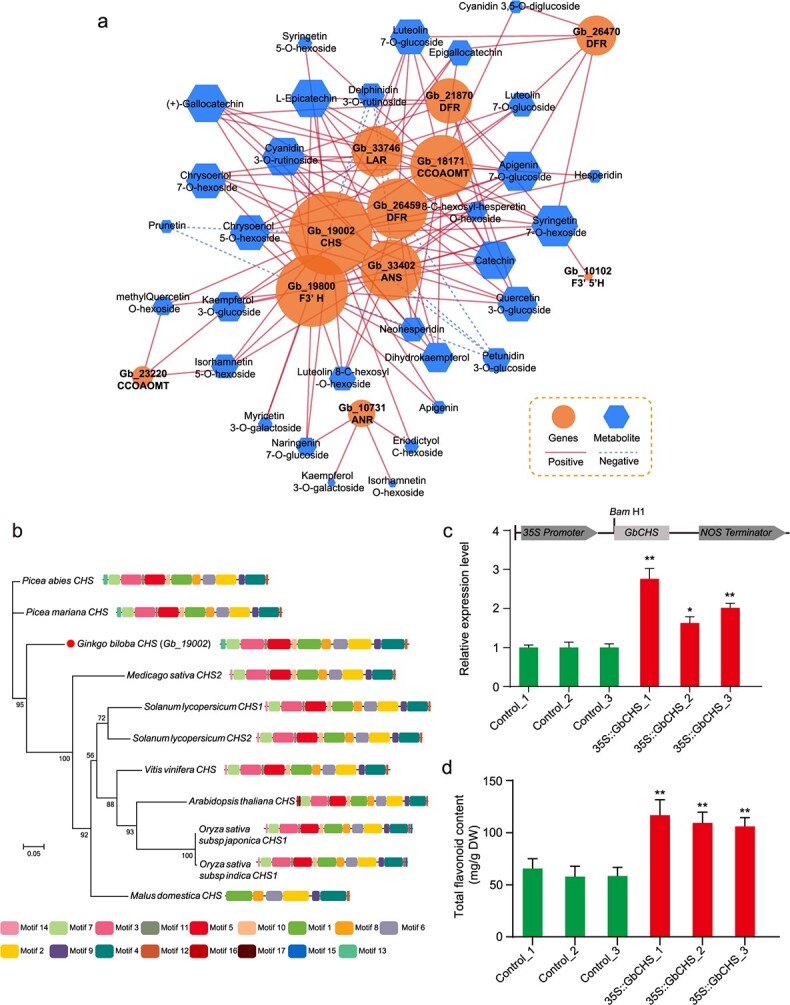
Association analysis and functional validation of transcriptomic and metabolic profiles related to flavonoid biosynthesis in rejuvenated leaves. a Correlation network of differentially expressed genes (DEGs) and metabolites. The correlations were estimated in accordance with an r2 value ≥0.9 and a p value <0.05, and the network was visualized using Cytoscape software (The Cytoscape Consortium, San Diego, CA, USA, version 3.5.1). The node size represents the number of differentially abundant metabolites associated with a particular DEG. ANR, anthocyanidin reductase; ANS, anthocyanidin synthase; CHI, chalcone isomerase; CHS, chalcone synthase; CCOMT, caffeoyl-CoA O-methyltransferase; DFR, dihydroflavonol 4-reductase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′5′-hydroxylase; FLS, flavonol synthase; FNS1, flavone synthase 1; LAR, leucoanthocyanidin reductase. b Phylogenetic tree analysis of CHS proteins from other species. c qRT–PCR-based confirmation of GbCHS-overexpressing transgenic G. biloba calli. d Total flavonoid contents of the transgenic calli and controls.
We further cloned the complete CDS of GbCHS (Gb_19002) from G. biloba leaves (Fig. S5). Phylogenetic analysis showed that GbCHS is closely related to CHS in gymnosperms such as Picea abies and Picea mariana (Fig. 6b). Amino acid sequence analysis indicated that GbCHS has many conserved motifs shared among gymnosperms and angiosperms. We constructed a pRI101-GbCHS plasmid in which the gene was driven by the cauliflower mosaic virus 35S promoter. Agrobacterium-mediated 35S::GbCHS transient transformation was performed in G. biloba calli, and the expression of GbCHS in three independent transformed calli was confirmed via qRT–PCR (Fig. 6c). In addition, the total flavonoid content significantly increased in transformed calli compared to nontransformed calli (controls) (Fig. 6d), further implicating CHS as being involved in flavonoid biosynthesis in G. biloba.
Changes in phytohormone levels in rejuvenated leaves
To explore the endogenous phytohormone changes in the rejuvenated leaves, we quantified the concentrations of four hormones, auxin (indole-3-acetic acid [IAA]), cytokinins (zeatin riboside [ZR]), GA3, and ABA, at three developmental stages. Compared to those of the controls (33.27 ± 3.78 ng/g in S1; 3.29 ± 1.32 ng/g in S5), the IAA contents of the truncation samples significantly increased at S1 (58.46 ± 6.85 ng/g) and S5 (6.89 ± 0.86 ng/g) (Fig. 7a); in contrast, the ABA content of the truncated plants decreased by 51.45% in S1 (to 55.69 ± 6.74 ng/g), but no statistically significant differences were observed for S3 and S5 (Fig. 7b). The ZR content of the truncated plants was also significantly lower than that of the control in S1, whereas the opposite tendency was observed in S3 and S5 (Fig. 7c). Interestingly, the GA3 content increased significantly across all three stages in the rejuvenated leaves (Fig. 7d), with average levels of 44.79 ng/g in S1, 33.19 ng/g in S3, and 27.26 ng/g in S5, compared to those of the controls (38.8 ng/g in S1, 27.41 ng/g in S3, and 21.46 ng/g in S5). Consistently, GAs composed 31.6% (6/19) of all phytohormone metabolites detected in the metabolome (Fig. 7e), of which four GAs (GA3, GA4, GA15, and GA20) presented higher contents in the rejuvenated leaves than in the controls (Fig. 7e, f).
Figure 7.
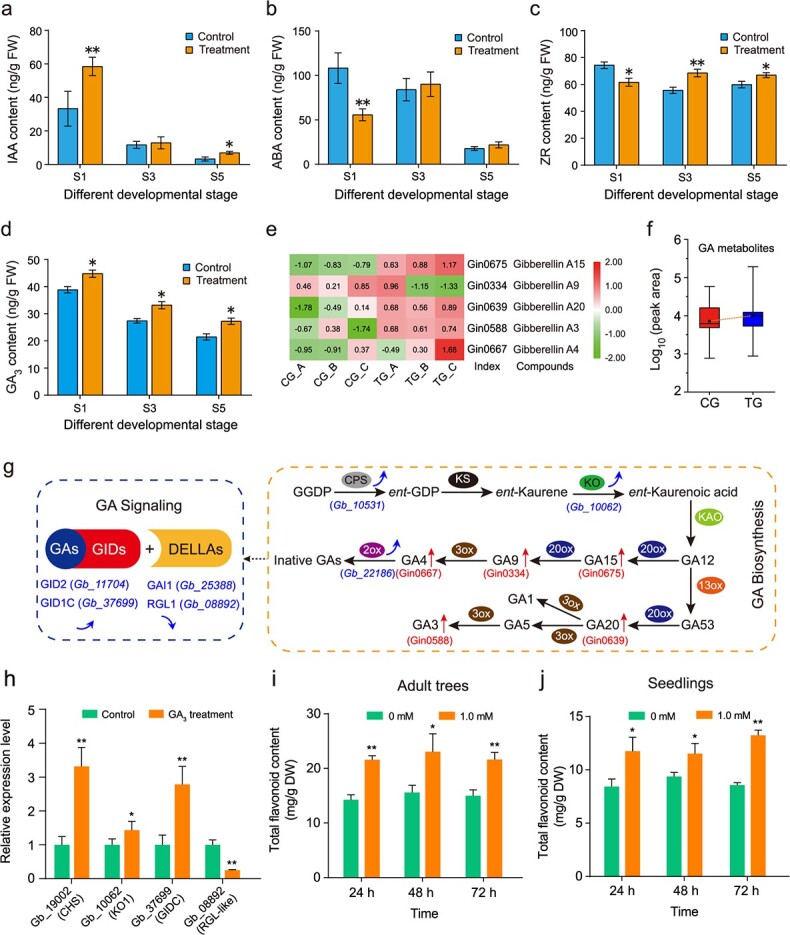
Changes in endogenous hormone levels, related gene expression levels, and leaf flavonoid contents. a–d Indole-3-acetic acid (IAA; a), abscisic acid (ABA; b), zeatin riboside (ZR; c), and gibberellic acid (GA3; d) contents were assayed in control and rejuvenated leaves at S1, S3 and S5. The data are the means of three biological replicates, and the error bars represent the SDs. FW, fresh weight. * and ** indicate significant differences from the controls at p < 0.05 and p < 0.01, respectively, as determined using Student’s t-test. e–f Increased contents of the various gibberellin (GA) metabolites in rejuvenated leaves. The data are the means and SDs of relative abundance levels (n = 3). g Genes and metabolites involved in GA biosynthesis and signaling. The blue arrows indicate expression changes (decrease or increase) of genes following the truncation treatments. qRT–PCR-based assessment of the genes in Fig. S6. The red arrows indicate the abundance changes of metabolites (red) following the truncation treatments. h qRT–PCR-based assessment of the expression of select genes involved in flavonoid biosynthesis (CHS), GA biosynthesis (KO1), and GA signaling (GID1C and RGL1) at 24 h after exogenous GA3 treatment (1.0 mM, adult trees). i–j Changes in the total flavonoid content of G. biloba leaves after exogenous GA3 treatments. Twenty-year-old adult trees (i) and 6-month-old Ginkgo saplings (j) were sprayed with 1.0 mM exogenous GA3. Leaf flavonoid contents were measured for 3 days. The data are the means and SDs (n ≥ 3 for each treatment). * and ** indicate significant differences from the controls at p < 0.05 and p < 0.01, respectively, as determined by ANOVA with post hoc tests.
We compared the expression changes of genes involved in GA biosynthesis (ENT-KAURENE OXIDASE [KO1], ENT-COPALYL DIPHOSPHATE SYNTHASE [CPS], and GIBBERELLIN 2-BETA-DIOXYGENASE 1 [GA2ox1]) and signaling (GIBBERELLIN-INSENSITIVE DWARF PROTEIN 2 [GID2], GIBBERELLIN INSENSITIVE DWARF 1C [GID1C], GIBBERELLIC ACID-INSENSITIVE MUTANT PROTEIN 1 [GAI1], and REPRESSOR of GIBBERELLIC ACID LIKE 1 [RGL1-like]) pathways between the truncated and control leaves of trees at all three stages (Fig. S6). The expression of the genes encoding KO1, CPS, and GA2ox1 significantly increased in the leaves of the truncated trees at S1 and S5. The expression levels of GID2 and GID1C, which encode receptors involved in GA signaling, were significantly higher in the truncation sample than in the controls across the three stages (S1, S3, and S5). In contrast, genes encoding the DELLA proteins GAI1 and RGL1, which are repressors of the GA signaling pathway, were expressed at significantly lower levels in the truncated trees than in the controls. The expression patterns of these genes, with the exception of CPS, generally exhibited similar patterns between the qRT–PCR data and RNA-seq data.
GA regulates GbCHS to promote flavonoid biosynthesis in rejuvenated leaves
We conducted exogenous GA treatments to further determine the role of GAs in response to truncation. We examined the expression of GA biosynthesis- and signaling-related genes in adult trees after 1.0 mM GA3 treatment. As expected, the expression levels of KO1 and GID1C were higher in the GA3 treatment than in the controls, whereas GAI1 and RGL1-like were downregulated in response to GA3 treatment (Fig. 7h). Notably, the flavonoid biosynthesis gene GbCHS was significantly upregulated in response to GA3. Under 1.0 mM GA3, the accumulation of total flavonoids significantly increased (Fig. 7i). Consistent with the findings in the adult trees, 1.0 mM GA3 significantly increased the total flavonoid content 1 day after treatment in the saplings (Fig. 7j). These results demonstrate that exogenous application of GA3 increases the flavonoid content by triggering the expression of key flavonoid biosynthesis-related genes.
Discussion
Trees grow in multiple directions through their apical and lateral meristems. Periodic growth over many years results in a complex pattern of varying physiological ages in different parts of the tree, including on different branches and even at different positions along the same branch [3]. Juvenile characteristics are often retained in a “cone of juvenility” at the center and base of a tree, whereas the more distant shoots are more mature [5, 9]; for example, in Acacia koa, pinnately compound leaves are produced at the base trunk (the juvenile phase), whereas phyllode leaves are produced from the top of the trunk (the adult phase) [3].
Regeneration is based on cell totipotency and pluripotency, via which organisms repair or replace themselves after damage or stress [44, 45]. Continuously pruning seedlings can maintain their juvenility or delay their maturation. In addition, when a tree is cut down, coppice shoots are produced from the stump and appear more juvenile than the shoots in the crown of mature trees do [2, 46]. Generally, rejuvenated shoots have higher photosynthetic rates and chlorophyll contents than do adult shoots [8]. Rejuvenated cuttings can also increase rooting capacity and improve root morphological variables [6, 45] and are therefore used in clonal forestry applications. In the present study, we truncated G. biloba trees to different heights and compared the morphological changes between the regenerated shoots and leaves. We found that after truncation, many sprouts were produced between the base of the trunk and the pruning position. The number of leaf lobes dramatically increased in these new shoots. In Ginkgo leaf fossils, more divided lobes were present in the leaves of Jurassic and Cretaceous species [47], whereas in extant G. biloba, leaves at the top of long shoots had more lobes than did leaves on short shoots [42]. Leaves from young G. biloba saplings also display the deep-lobed leaf phenotype [43]. Therefore, the more deeply divided leaves observed in the truncated G. biloba trees may reflect the ancestor’s morphological characteristics, particularly because the juvenile characteristics of young saplings may reflect primitive traits.
Several plant developmental characteristics, including leaf structure and morphology, are important determinants of photosynthetic efficiency and the overall performance of plants [48]. The optimization of these developmental traits has considerable potential for increasing biomass and yield; for instance, leaves of an optimal shape and size promote more efficient light harvesting, leading to a faster growth rate and increased biomass [49]. Leaf biomass is largely dependent on the photosynthetic capacity of the plant as a whole, and enhancing photosynthesis at the level of a single leaf would increase yield [50]. In this study, the Ginkgo trees exhibited many enhanced developmental features in association with the regenerated branches, including significantly increased shoot growth vigor, leaf size, leaf thickness, and leaf fresh/DW with delayed senescence; this enhanced leaf biomass. Although the leaf yield of the truncated trees was lower than that of controls during the first year, a large increase in yield occurred in the second year; therefore, combined with high-density planting, the leaf yield can be dramatically increased. In addition, a dwarf (truncated) tree form facilitates the collection of leaves by manual or mechanical harvesting. Thus, regeneration by truncation is effective for improving leaf biomass and yield in G. biloba.
Endogenous phytohormones play important roles in restoring the juvenile features of trees [2, 6]. Cytokinins, auxins, and GAs can induce rejuvenation and maintain trees in a juvenile state [10, 51–53]. The IAA/ABA levels also reflect the extent of the juvenile phenotype in vitro and the rooting abilities of young stems [4, 6, 54]. Pretreatment of shoots with GA3 improves adventitious rooting of cuttings, demonstrating the important role of this phytohormone in rejuvenation [10]. In this study, the contents of IAA at S1 and S5 were significantly higher in the truncated trees than in the controls, whereas the ZR content was higher at S3 and S5, indicating that these two hormones may be involved in the rejuvenation process. Furthermore, the GA3 content was significantly higher in the rejuvenated samples than in the controls at all three developmental stages. Our metabolomic analysis revealed that five (GA3, GA4, GA9, GA15, and GA20) of the six GAs studied were more abundant in the rejuvenated leaves than in the control leaves. In addition, the expression levels of GA biosynthesis-related genes (KO1, CPS, and GA2ox1) and signaling-related genes (GID2 and GID1C) were upregulated in the rejuvenated leaves, whereas the DELLA protein-encoding genes GAI and RGL1 were downregulated. These results suggest that truncation treatment activates GA biosynthesis and signaling by inducing the transcription of associated genes, which consequently contributes to the rejuvenation of G. biloba trees.
Flavonoids are among the most important secondary metabolites in plants, and their accumulation can be affected by age and the juvenility–maturity processes that are ongoing throughout the lifespan of the plant [27, 32, 55]. In Arabidopsis, the flavonol glycoside content in the cotyledons gradually decreased as the leaves matured [56]. In contrast to herbaceous plants, trees show a more complex pattern of flavonoid accumulation throughout their lifespans. Previous studies demonstrated that the contents of total flavonol glycosides in G. biloba leaves generally decreased with tree age [41]. Here, the rejuvenated G. biloba leaves produced following the truncation treatments contained at least 14% more total flavonoids than the control leaves did, indicating a significant correlation between rejuvenation and flavonoid accumulation. Furthermore, in combination with widely targeted metabolomics, we found that more than 87% of the differentially abundant flavonoids, including flavonol glycosides (quercetin glycosides, kaempferol glycosides, isorhamnetin glycosides) and flavones (apigenin, luteolin glycosides), significantly increased in the truncated plants. These results suggested that rejuvenation by truncation significantly promotes the accumulation of flavonoids in rejuvenated G. biloba trees.
The transcript abundance of genes involved in flavonoid biosynthesis could be coordinately regulated by the developmental transition from juvenility to maturity [32]. SPLs targeted by miR156 affect a broad range of developmental processes in Arabidopsis, including activating the vegetative phase transition and inducing flowering [21]. Phase transitions are closely correlated with secondary metabolism in plants. During the plant transition to flowing, SPL9 negatively regulates anthocyanin levels by interfering with the MYB-bHLH-WD40 transcription complex and the expression of the anthocyanin biosynthesis-related DFR gene [32]. In addition, SPL9 and its regulator, miR156, control anthocyanin metabolism in an age-dependent manner [57]. Our RNA-seq and qRT–PCR results indicated that truncation significantly increased the expression levels of flavonoid biosynthesis genes, including CHS, FLS, DFR, and F3’H. In particular, compared with that in the controls, CHS in the leaves of truncated trees was consistently expressed at a 2.5-fold higher level, suggesting that they contribute to the accumulation of flavonols and flavones in rejuvenating trees. Further cloning and overexpression analysis of GbCHS revealed a marked increase in total flavonoid contents in those G. biloba calli compared to controls, confirming that GbCHS functions in flavonoid biosynthesis. In addition, we identified three SPL genes—SPL3, SPL8, and SPL12—that displayed decreased expression levels at S1 but significantly increased levels at S5 in the truncated trees, thus showing no correlation with rejuvenation in G. biloba.
Development-associated phytohormones also play important roles in regulating flavonoid biosynthesis, with changes in the abundance of auxin, cytokinins, GAs, or their crosstalk coordinating the accumulation of or decrease in flavonoids in plant organs or tissues [58, 59]. In Arabidopsis, auxin and ethylene induce flavonol accumulation via distinct signaling networks [60], whereas GAs negatively regulate flavonol biosynthesis and modulate root growth [61]. Exogenous application of GA3 significantly increased the contents of flavonoids in hairy roots and regenerated plants of Saussurea involucrata [62], and GA3 treatment of grapevine (Vitis labrusca × Vitis vinifera) markedly upregulated the expression levels of CHS, F3H, and PAL (PHENYLALANINE AMMONIA-LYASE), which are involved in flavonoid biosynthesis [63]. To assess the effects of GA3 on flavonoid biosynthesis, we performed exogenous GA3 treatments. GbCHS expression and total flavonoid contents were increased in the leaves of saplings and adult trees. We conclude that the increase in GA following the truncation treatment promotes GbCHS expression in G. biloba, both of which promote the flavonoid accumulation observed in rejuvenated leaves.
Taken together, our findings suggest a viable technique to improve the biomass and flavonoid content of Ginkgo leaves. Effective rejuvenation techniques are advantageous for maintaining juvenility and optimizing the growth of plants [9]. When plants are subjected to rejuvenation cuttings, the growth rate, leaf number, and biomass production significantly increase, indicating enhanced developmental traits [64]. The truncation treatment described here represents a promising rejuvenation technique for increasing leaf size, fresh weight (FW), photosynthetic capacity, and leaf biomass and delaying senescence in G. biloba. In addition, increasing the production of bioactive flavonoids in medicinal plant species is desirable. We found that the flavonoid biosynthesis gene CHS was highly upregulated in G. biloba following truncation treatment and that the contents of flavonol glycosides, including quercetin, kaempferol, and isorhamnetin glycosides, correspondingly increased. Exogenous GA treatment of G. biloba leaves promoted GbCHS expression and flavonoid accumulation.
In summary, we demonstrated that rejuvenation by truncation enhanced leaf production in G. biloba (Fig. 8). The enhanced biomass and flavonoid contents suggest that our approach can be applied to a variety of relevant medicinal plant species with regeneration capacity to increase yields and the abundance of target compounds. The enhancement of flavonoid synthesis after rejuvenation in other plant species is therefore worthy of investigation.
Figure 8.
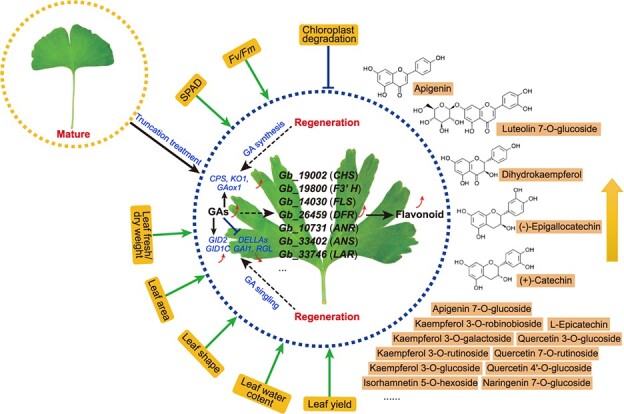
Model of the physiological and molecular mechanisms underlying flavonoid accumulation in rejuvenated leaves of G. biloba.
Materials and methods
Plant materials, growth conditions, and truncation treatments
Five-year-old G. biloba plants (Fozhi cultivar) were grown at a leaf harvesting ginkgo plantation located in Sihu, Pizhou city, Jiangsu Province, China (34.35°N, 117.59°E), in well-drained and moderately fertile soil. In this plantation, there were three sample plots, each of which was divided into truncation and control areas. Before truncation treatments, the trees of each sample plot were similar in terms of size, height, and growth status. The truncation treatments were applied on January 15, 2016. A total of 180 well-established trees from the three sample plots were used in the experiment, with 30 trees each being truncated to stem heights of 10, 35, 60, 85, and 110 cm above the ground; the remaining 30 trees were used as the controls. After a period of growth, the leaves were sampled continuously from the leaf expansion stage (April 2016) to the leaf-harvesting stage (September 2016), particularly at S1 (April 26), S2 (June 5), S3 (July 11), S4 (August 15), and S5 (September 25, the leaf-harvesting period). For each treatment, the leaves were collected separately from the middle-upper long branches (approximately 70% of the total height above the ground, adjacent to the cutting position) and lower long branches (approximately 30% of the total height above the ground, close to the base of trunk) of the experimental trees. These samples were used for morphological and anatomical observations, physiological measurements, and transcriptome sequencing. For the metabolomic and RNA-seq samples, leaves from the middle-upper branches of three individual trees were used as biological replicates for each treatment. All samples intended for molecular and metabolic analyses were immediately frozen in liquid nitrogen and stored at −80°C until used.
Physiological measurements and leaf yield estimation
Leaves at the leaf-harvesting stage were used for determination of lobe number, thickness, area, FW/DW, and water content. Approximately 30–50 fresh leaves per treatment were randomly collected from 10–15 plants and immediately placed into plastic self-sealing bags within a foam box that contained ice. Leaf lobes with depths below 20% of the leaf blade were not calculated. The leaf FW was measured using an analytical balance (MS205DU; Mettler Toledo, OH, USA), and the leaves were dried at 70°C to a constant DW. The leaf water content (%) was calculated according to the formula (FW–DW) ÷ FW × 100%. The leaf surfaces were imaged using a digital camera, and the leaf area was determined using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The physiological measurements were performed on at least 10 leaves per treatment, with three biological replicates. Additionally, all fresh leaves from the 30 plants under each truncation treatment were separately harvested on September 25, 2016, and September 20, 2017, and weighed for leaf biomass estimation. The leaf yield per hectare was estimated using the following formula: leaf yield (kg/hectare) = number of plants per hectare × leaf biomass per plant. In addition, the lengths of the new branches that sprouted near the cutting position and trunk base (at approximately 30% of the full trunk height) were measured and recorded at each stage.
Assays of chlorophyll content and photosynthetic parameters
In each treatment group, the soil–plant analysis development (SPAD) values of leaves from the middle-upper parts of the trees were measured at the leaf-harvesting stage using a SPAD-502 Plus chlorophyll meter (Konica Minolta, Osaka, Japan). Three SPAD readings were uniformly obtained at the middle region of each leaf blade. Due to the unique fan shape of G. biloba leaf blades, it is easy to determine the location of the middle region of the leaf blade, which ensures that the measurement position of each leaf blade is identical. Photosynthetic parameter measurements were conducted between 12:00 and 13:00 h on sunny days, with data collected from the plants at different developmental stages. Mature leaves borne on long branches located in the middle-upper region of the plants were selected. Chlorophyll fluorescence parameters were measured using an LI-6400XT portable photosynthesis system with an integrated fluorescence fluorometer (LI-6400-40 leaf chamber fluorometer; LI-COR, Lincoln, NE, USA). After dark adaptation of leaves for 20 min, the maximal fluorescence (Fm), minimal fluorescence (Fo), and maximum quantum efficiency values of photosystem II (Fv/Fm) were recorded. For the SPAD value and chlorophyll fluorescence parameter measurements, at least five individual trees under each treatment were randomly selected, with at least eight measurements taken per individual tree at each stage.
Transmission electron microscopy
Fresh tissue samples (5 × 5 mm; 85 cm truncation treatment) at S5 were harvested from the middle part of each leaf along the central vein. The samples were processed and embedded in Spurr’s resin, as described previously [65]. Thin (80-nm) sections were cut and stained for observations under a transmission electron microscope (JEOL, Ltd., Tokyo, Japan).
Measurements of flavonoid and terpenoid contents
G. biloba leaves at S5 are commonly used for commercial leaf harvesting [24]; therefore, the terpene and flavonoid contents were measured in leaves at this stage under different treatments by high-performance liquid chromatography-mass spectrometry (HPLC–MS). For the terpene lactones, 1.5 g of ginkgo leaves was dried in an oven (60°C) and ground to a powder to extract the terpene lactones. The extraction of terpene lactones was performed according to the method of the Chinese Pharmacopoeia Commission. An Atlantis T3 column (4.6 × 150 mm, 4 μm) was used to separate terpene lactones. Analysis of the terpene lactones was conducted using a Waters 2424 HPLC system (Waters, Milford, MA, USA) equipped with an Alltech ELSD 2000 evaporative light-scattering detector (Alltech, Deerfield, IL, USA). The mobile phase was composed of tetrahydrofuran, methanol, and water at a ratio of 10:25:65 (v/v/v). The mobile phase and ELSD parameters were as follows: a 0.35 mL/min flow rate of tetrahydrofuran-methanol; a 0.65 mL/min flow rate of water; a 100°C ELSD drift tube temperature; and a 35°C column temperature. The total content (mg/g) of terpene lactones was determined as the sum of the ginkgolide A, ginkgolide B, ginkgolide C, and bilobalide contents. All standards used for confirmation of terpenoids were obtained from Shanghai Yuanye Bio-Technology Company (Shanghai, China). The total flavonoid content of a 0.02 g ginkgo leaf sample was also determined in accordance with the protocol of a plant flavonoid test kit (Suzhou Comin Biotechnology Co., Ltd., Suzhou, China). Three independent biological replicates were included for each experiment.
Endogenous hormone measurements
G. biloba leaves of trees under the 85-cm truncation treatment at developmental stages S1, S3, and S5 were analyzed for IAA, ZR, ABA, and GA3 concentrations. The sample preparation, extraction, and purification of ABA, IAA, and GA3 were performed as described by Pan et al. [66] and Wang et al. [67]. Briefly, 10 μL of sample solution was injected onto an Agilent Eclipse Plus C18 column (150 × 2.1 mm, 5 μm) for analysis using an Agilent 6460 HPLC–electrospray ionization–tandem mass spectrometry (ESI–MS/MS) system. The HPLC–ESI–MS/MS and multiple reaction monitoring (MRM) conditions and settings are the same as those described above for the measurement of terpene lactones, with the exception of the elution program. The elution program used to determine the endogenous hormone contents was as follows: 0–5 min, 10% A (acetonitrile) + 90% B (0.1% formic acid solution); 5–10 min, 70% A + 30% B; 10–13 min, 90% A + 10% B; and 13–20 min, 10% A + 90% B. The ZR content could not be quantified by HPLC-ESI-MS/MS; therefore, an enzyme-linked immunosorbent assay (ELISA) kit (Shanghai Qiaodu Biological Technology Co., Ltd., Shanghai, China) was used to quantify the ZR level. The standards used for confirmation of hormones were purchased from Shanghai Yuanye Bio-Technology Company. Three biological replicates were included for each set of experiments.
Metabolite identification and quantification by ultra-performance liquid chromatography–electrospray ionization–tandem mass spectrometry
Leaf samples from trees subjected to the 85-cm truncation treatment and control trees were collected during the leaf-harvesting period for metabolite identification and quantification. Three biological replicates were independently analyzed via an ultra-performance liquid chromatography–ESI)–quadrupole time of flight (QTOF)–MS/MS system (UPLC: UFLC CBM30A system, Shimadzu, Kyoto, Japan. MS: Applied Biosystems 4500 QTRAP, Waltham, MA, USA). The effluent was connected to an ESI-triple quadrupole-linear ion trap (QTRAP)–MS/MS system. The ESI source parameters were as follows: positive and negative ionization modes; a 5.5 kV ion spray voltage; and a 500°C source temperature. The monitoring mode was set to MRM.
Metabolite identification and quantification were performed in accordance with the method described by Chen et al. [68]. Metabolite identification was dependent on the primary and secondary MS data annotated against an in-house database and publicly available metabolite databases. The metabolites were quantified using MRM. PCA and OPLS–DA were used to comprehensively compare the truncation treatment and control samples. Significantly differentially abundant metabolites were identified according to a P value (t-test) of <0.05 and a variable importance in projection value of ≥1. The differentially abundant metabolites were mapped to KEGG metabolic pathways for pathway and enrichment analyses.
RNA-seq
The leaves of the truncated and control trees were collected at S5 for RNA extraction. Total RNA was isolated from three biological replicates using a MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. cDNA libraries were subsequently constructed as described previously [69] and sequenced on a HiSeq 4000 platform (Illumina, San Diego, CA, USA).
The generated raw reads were filtered to remove adaptors, low-quality sequences with unknown nucleotides (N), and reads with more than 20% low-quality bases (base quality <10). The filtered reads were aligned to the G. biloba genome (http://gigadb.org/dataset/100209) using TopHat 2 [70]. The gene expression level was quantified according to the fragments per kilobase of transcript per million mapped reads (FPKM). Genes that were differentially expressed between the treatment and control groups were identified using the edgeR package [71] on the basis of a P value ≤0.05 and a log2(ratio) ≥ 1. All DEGs were automatically annotated, categorized automatically using Blast2GO and mapped to KEGG pathways. Heatmaps of gene expression were constructed using TBtools [72].
qRT–PCR
To confirm the expression changes of a subset of candidate genes, several genes associated with rejuvenation, secondary metabolites, GA biosynthesis, and GA signaling from trees at S1, S3, and S5 were selected and analyzed via qRT–PCR. Gene-specific primers were designed using Primer 5.0 (Table S2), and G. bilobaGAPDH expression was used as an internal control. qRT–PCR was performed as described previously [69]. Relative gene expression levels were calculated using the 2−ΔΔCt method. All reactions were repeated for three biological replicates.
Exogenous hormone treatments
Six-month-old G. biloba saplings were cultivated from seeds collected from a single plant. The saplings were grown in a growth chamber at 25/20°C (day/night) with a 16 h light/8 h dark photoperiod and ~ 65% humidity. Twenty-year-old Ginkgo trees growing in the leaf-harvesting plantation were also used.
For the 20-year-old trees (cultivar Fozhi), branches (~50 cm) with leaves growing at the same height and position (sun-exposed slope) were sampled from three individual trees and precultured in water for 6 h to minimize individual differences. These branches (five replicates per group) were treated with 1.0 mM GA3 (three groups); water was used as the control treatment. The leaves of the branches were sprayed with 1.0 mM GA3 (plus 0.02% Tween-20). At 24, 48, and 72 h after treatment, the leaves subjected to each treatment were separately collected. The total flavonoid contents of the leaves were determined as described above. The expression levels of GA biosynthesis- and signaling-related genes and GbCHS were examined in the leaves of adult trees (1.0 mM, 24 h after spraying) via qRT–PCR.
For the 1-year-old saplings (12 individuals), the aboveground parts of plants were sprayed with 1.0 mM GA3 (plus 0.02% Tween-20) during the day (six plants per treatment). Water including 0.02% Tween-20 was used as the control treatment. At 24, 48, and 72 h after spraying, the leaves were separately sampled and frozen in liquid nitrogen.
Sequence and phylogenetic analyses of GbCHS
The nucleotide and amino acid sequences of GbCHS (Gb_19002) were obtained from G. biloba genomic information. The protein sequences from multiple species (A. thaliana, Oryza sativa, V. vinifera, Malus domestica, Solanum lycopersicum, Medicago sativa, Picea abies, P. mariana) were downloaded from the SwissProt database. Conserved motif sequences were analyzed using MEME Suite. Multiple sequence alignments of CHS protein sequences were performed by ClustalX, and a phylogenetic tree was constructed using IQ-TREE (v.1.6.9), with the maximum-likelihood method.
Vector construction and transformation
Gene-specific primers for GbCHS were designed using Primer Premier 5.0. The GbCHS coding region without the stop codon (TGA) was amplified from the cDNA of G. biloba leaves. The PCR products were inserted into the intermediate vector pMD-19 T (TaKaRa), and a pMD-19 T-GbCHS plasmid was used to isolate GbCHS for homologous recombination. The plant expression vector pRI101-GFP was digested with the Bam HI site. The GbCHS fragment was inserted into pRI101-GFP to generate 35S::GbCHS-GFP via a Exnase II Cloning Kit (Vazyme Biotech, Nanjing, China). After sequencing validation, the recombinant plasmid pRI101-GbCHS-GFP was obtained; an empty vector was used as a control. To validate the function of GbCHS, the constructs and empty vector (control) were introduced into induced G. biloba leaf calli via Agrobacterium-mediated transient transformation. Approximately four days after transformation, the expression of GbCHS and other GA-related genes in the G. biloba calli (~30 days of cultivation on growth media) was assessed. Moreover, the total flavonoid content of transgenic G. biloba calli and the controls was determined as described above.
Statistical analysis
The phenotypic and physiological data (chlorophyll, photosynthesis, metabolite, and hormone measurements) and qRT–PCR data are presented as the means ± standard deviations (SDs) of at least three independent experiments. The statistical significance of differences between the treatment and control conditions was assessed by two-sided Student’s t-tests and ANOVA with post hoc tests. P values <0.05 and < 0.01 were regarded as statistically significant and highly significant, respectively.
Acknowledgments
This work was financially supported by National Key R&D Program of China (Grant No. 2017YFD0600701), the National Natural Science Foundation of China (Nos. 31971686 and 32072610), the Six Talent Peaks Project in Jiangsu Province (No. NY-090), and the Yangzhou University International Academic Exchange Fund. We thank the Genedenovo Biotechnology Co., Ltd., Guangzhou, China, for conducting the UPLC-ESI-MS/MS metabolomics work, RNA-seq and technical assistance. We thank Mr. Mingxian Wang and Jiangsu Dadunzi at Ginkgo Biotechnology Co., Ltd., China, for providing us with the ginkgo plantation used in this study.
Author contributions
J.L., L.W., B.J., and W.L. carried out the design of the study. Z.L. and L.Z. performed the experimental work, analyzed the data, and drafted the manuscript. J.L., N.S., L.W., S.L., Q.W. and W.Y. participated in the sample collection and data analysis. K.H. reviewed and revised the manuscript. All authors have read and approved the final manuscript.
Data availability statements
The sequencing and expression data are available at the NCBI Gene Expression Omnibus (GEO) under accession number GSE149736.
Conflicts of interest statement
The authors declare that they have no conflicts of interest.
Supplementary data
Supplementary data is available at Horticulture Research Journal online.
Supplementary Material
References
- 1. Greenwood MS. Juvenility and maturation in conifers, current concepts. Tree Physiol. 1995;15:433–8. [DOI] [PubMed] [Google Scholar]
- 2. Pei D, Gu R. A review on the rejuvenation of mature trees. Chine Bull Bot. 2005;22:753–60. [Google Scholar]
- 3. Poethig RS. The past, present, and future of vegetative phase change. Plant Physiol. 2010;154:541–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barthelemy D, Caraglio Y. Plant architecture: a dynamic, multilevel and comprehensive approach to plant form, structure and ontogeny. Ann Bot. 2007;99:375–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heide OM. Juvenility, maturation and rejuvenation in plants, adventitious bud formation as a novel rejuvenation process. J Hortic Sci Biotechnol. 2019;94:2–11. [Google Scholar]
- 6. Zhang Z, Sun Y, Li Y. Plant rejuvenation: from phenotypes to mechanisms. Plant Cell Rep. 2020;39:1249–62. [DOI] [PubMed] [Google Scholar]
- 7. Wang JW, Park MY, Wang L-Jet al. . miRNA control of vegetative phase change in trees. PLoS Genet. 2011;7:e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wendling I, Trueman SJ, Xavier A. Maturation and related aspects in clonal forestry—part I: concepts, regulation and consequences of phase change. New For. 2014b;45:449–71. [Google Scholar]
- 9. Wendling I, Trueman SJ, Xavier A. Maturation and related aspects in clonal forestry-part II: reinvigoration, rejuvenation and juvenility maintenance. New For. 2014a;45:473–86. [Google Scholar]
- 10. Ford YY, Taylor JM, Blake PSet al. . GibberellinA3 stimulates adventitious rooting of cuttings from cherry (Prunus avium). Plant Growth Regul. 2002;37:127–33. [Google Scholar]
- 11. Gordon SP, Heisler MG, Venugopala Get al. . Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development. 2007;134:3539–48. [DOI] [PubMed] [Google Scholar]
- 12. Sun T. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–45. [DOI] [PubMed] [Google Scholar]
- 13. Hackett WP. Juvenility, maturation and rejuvenation in woody plants. Hortic Rev. 1985;7:109–55. [Google Scholar]
- 14. Evans MM, Poethig RS. Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol. 1995;108:475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manuela D, Xu M. Juvenile leaves or adult leaves: determinants for vegetative phase change in flowering plants. Int J Mol Sci. 2020;21:9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Telfer A, Poethig RS. HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development. 1998;125:1889–98. [DOI] [PubMed] [Google Scholar]
- 17. Tanaka N. Gibberellin is not a regulator of miR156 in rice juvenile-adult phase change. Rice. 2012;5:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lauter N, Kampani A, Carlson SRet al. . microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci U S A. 2005;102:9412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang T, Lian H, Tang Het al. . An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell. 2015;27:349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye BB, Zhang K, Wang JW. The role of miR156 in rejuvenation in Arabidopsis thaliana. J Integr Plant Biol. 2020;62:550–5. [DOI] [PubMed] [Google Scholar]
- 21. Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–49. [DOI] [PubMed] [Google Scholar]
- 22. Yu S, Galvao VC, Zhang Y-Cet al. . Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell. 2012;24:3320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahsan MU, Hayward A, Irihimovitch Vet al. . Juvenility and vegetative phase transition in tropical/subtropical tree crops. Front Plant Sci. 2019;10:729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang L, Xu M, Koo Yet al. . Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. elife. 2013;2:e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu S, Cao L, Zhou C-Met al. . Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. elife. 2013;2:e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ponnu J, Schlereth A, Zacharaki Vet al. . The Trehalose 6-phosphate pathway impacts vegetative phase change in Arabidopsis thaliana. Plant J. 2020;104:768–80. [DOI] [PubMed] [Google Scholar]
- 27. Buer CS, Imin N, Djordjevic MA. Flavonoids: new roles for old molecules. J Integr Plant Biol. 2010;52:98–111. [DOI] [PubMed] [Google Scholar]
- 28. Feller A, Machemer K, Braun ELet al. . Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. [DOI] [PubMed] [Google Scholar]
- 29. Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–85. [DOI] [PubMed] [Google Scholar]
- 30. Yu Z-X, Wang L-J, Zhao Bet al. . Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol Plant. 2015;8:98–110. [DOI] [PubMed] [Google Scholar]
- 31. Mao Y-B, Liu Y-Q, Chen D-Yet al. . Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat Commun. 2017;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gou J, Felippes FF, Liu Cet al. . Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao Y-P, Fan G, Yin P-Pet al. . Resequencing 545 ginkgo genomes across the world reveals the evolutionary history of the living fossil. Nat Commun. 2019;10:4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu H, Wang X, Wang Get al. . The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat Plants. 2021;7:748–56. [DOI] [PubMed] [Google Scholar]
- 35. Lu J, Xu Y, Meng Zet al. . Integration of morphological, physiological and multi-omics analysis reveals the optimal planting density improving leaf yield and active compound accumulation in Ginkgo biloba. Ind Crop Prod. 2021;172:114055. [Google Scholar]
- 36. Zhang H-F, Huang L-B, Zhong Y-Bet al. . An overview of systematic reviews of Ginkgo biloba extracts for mild cognitive impairment and dementia. Front Aging Neurosci. 2016;8:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zuo W, Yan F, Zhang Bet al. . Advances in the studies of Ginkgo biloba leaves extract on aging-related diseases. Aging Dis. 2017;8:812–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shu Z, Shar AH, Shahen Met al. . Pharmacological uses of Ginkgo biloba extracts for cardiovascular disease and coronary heart diseases. Int J Pharmacol. 2019;15:1–9. [Google Scholar]
- 39. Singh SK, Srivastav S, Castellani RJet al. . Neuroprotective and antioxidant effect of Ginkgo biloba extract against AD and other neurological disorders. Neurotherapeutics. 2019;16:666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Momtazi-Borojeni AA, Katsiki N, Pirro Met al. . Dietary natural products as emerging lipoprotein(a)-lowering agents. J Cell Physiol. 2019;234:12581–94. [DOI] [PubMed] [Google Scholar]
- 41. Yao X, Shang E, Zhou Get al. . Comparative characterization of total flavonol glycosides and terpene lactones at different ages, from different cultivation sources and genders of Ginkgo biloba leaves. Int J Mol Sci. 2012;13:10305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leigh A, Zwieniecki MA, Rockwell FEet al. . Structural and hydraulic correlates of heterophylly in Ginkgo biloba. New Phytol. 2011;189:459–70. [DOI] [PubMed] [Google Scholar]
- 43. Bauer K, Grauvogel-Stamm L, Kustatscher Eet al. . Fossil ginkgophyte seedlings from the Triassic of France resemble modern Ginkgo biloba. BMC Evol Biol. 2013;13:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ikeuchi M, Ogawa Y, Iwase Aet al. . Plant regeneration: cellular origins and molecular mechanisms. Development. 2016;143:1442–51. [DOI] [PubMed] [Google Scholar]
- 45. Xu Z, et al. Plant cell totipotency and regeneration. Sci Sin Vitae. 2019;49:1282–300. [Google Scholar]
- 46. Pijut PM, Woeste KE, Michler CH. Promotion of adventitious root formation of difficult-to-root hardwood tree species. Hortic Rev. 2011;38:213–51. [Google Scholar]
- 47. Zhou Z, Zheng S. Palaeobiology: the missing link in Ginkgo evolution. Nature. 2003;423:821–2. [DOI] [PubMed] [Google Scholar]
- 48. Mathan J, Bhattacharya J, Ranjan A. Enhancing crop yield by optimizing plant developmental features. Development. 2016;143:3283–94. [DOI] [PubMed] [Google Scholar]
- 49. Meyer RS, Purugganan MD. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet. 2013;14:840–52. [DOI] [PubMed] [Google Scholar]
- 50. Long SP, Zhu X, Naidu SLet al. . Can improvement in photosynthesis increase crop yields. Plant Cell Environ. 2006;29:315–30. [DOI] [PubMed] [Google Scholar]
- 51. Zhang T-Q, Lian H, Zhou C-Met al. . Two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell. 2017;29:1073–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang J, Tian C, Zhang Cet al. . Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell. 2017;29:1373–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu H, Gao Y, Song Xet al. . A novel rejuvenation approach to induce endohormones and improve rhizogenesis in mature Juglans tree. Plant Methods. 2018;14:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guan L, Murphy AS, Peer WAet al. . Physiological and molecular regulation of adventitious root formation. Crit Rev Plant Sci. 2015;34:506–21. [Google Scholar]
- 55. Lucas WJ. Plant vascular biology and agriculture. J Integr Plant Biol. 2010;52:4–7. [DOI] [PubMed] [Google Scholar]
- 56. Peer KR, Greenwood MS. Maturation, topophysis and other factors in relation to rooting in Larix. Tree Physiol. 2001;21:267–72. [DOI] [PubMed] [Google Scholar]
- 57. Yang C-Q, Fang X, Wu X-Met al. . Transcriptional regulation of plant secondary metabolism. J Integr Plant Biol. 2012;54:703–12. [DOI] [PubMed] [Google Scholar]
- 58. Loreti E, Povero G, Novi Get al. . Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–16. [DOI] [PubMed] [Google Scholar]
- 59. Ji X-H, Wang Y-T, Zhang Ret al. . Effect of auxin, cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Organ Cult. 2015;120:325–37. [Google Scholar]
- 60. Lewis DR, Ramirez MV, Miller NDet al. . Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 2011;156:144–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tan H, Man C, Xie Yet al. . A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis. Mol Plant. 2019;12:521–37. [DOI] [PubMed] [Google Scholar]
- 62. Qiao X, Jiang S, Li Xet al. . Effects of phytohormones on plant regeneration and production of flavonoids in transgenic Saussurea involucrata hairy roots. Chine J Biotech. 2011;27:69–75. [PubMed] [Google Scholar]
- 63. Cheng C, Jiao C, Singer SDet al. . Gibberellin-induced changes in the transcriptome of grapevine (Vitis labrusca × V. vinifera) cv. Kyoho flowers. Genomics. 2015;16:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Greenwood MS, Day ME, Schatz J. Separating the effects of tree size and meristem maturation on shoot development of grafted scions of red spruce (Picea rubens Sarg.). Tree Physiol. 2010;30:459–68. [DOI] [PubMed] [Google Scholar]
- 65. Lu Z, Xu J, Li Wet al. . Transcriptomic analysis reveals mechanisms of sterile and fertile flower differentiation and development in Viburnum macrocephalum f. keteleeri. Front Plant Sci. 2017;8:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat Protoc. 2010;5:986–92. [DOI] [PubMed] [Google Scholar]
- 67. Wang L, Cui J, Jin Bet al. . Multifeature analyses of vascular cambial cells reveal longevity mechanisms in old Ginkgo biloba trees. Proc Natl Acad Sci U S A. 2020;117:2201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen W, Gong L, Guo Zet al. . A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites, application in the study of rice metabolomics. Mol Plant. 2013;6:1769–80. [DOI] [PubMed] [Google Scholar]
- 69. Li W-X, Yang S-B, Lu Zet al. . Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic Res. 2018;5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim D, Pertea G, Trapnell Cet al. . TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen C, Chen H, Zhang Yet al. . TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing and expression data are available at the NCBI Gene Expression Omnibus (GEO) under accession number GSE149736.