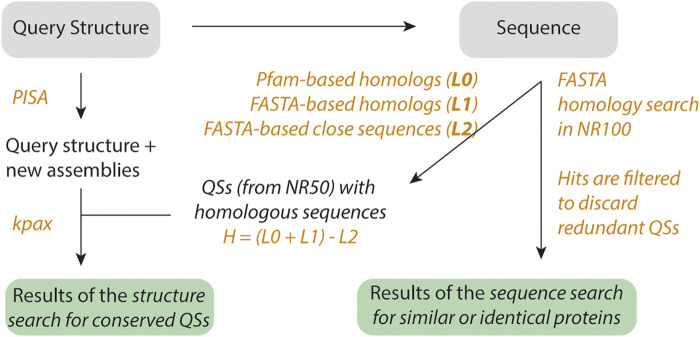
FIGURE 1.
Workflow of QSalignWeb. The user submits a query structure. Additional assemblies are identified using PISA. The resulting assemblies are each superposed with candidate QSs. Because structure superposition is computationally expensive, we only superpose QSs that exhibit the same number of subunits and show sequence homology. Homologs are identified based on two searches: A sequence similarity search with FASTA yields list L1, and a PFAM domain architecture similarity search yields list L0. We take the union of these two lists, and discard very close homologs (list L2, sequence identity > 80%). The structure superposition and inference of physiological relevance is carried out as described previously (Dey et al., 2018). On top of the results of the QS superposition, we also display a table of non-redundant QSs that share sequence similarity with the query.