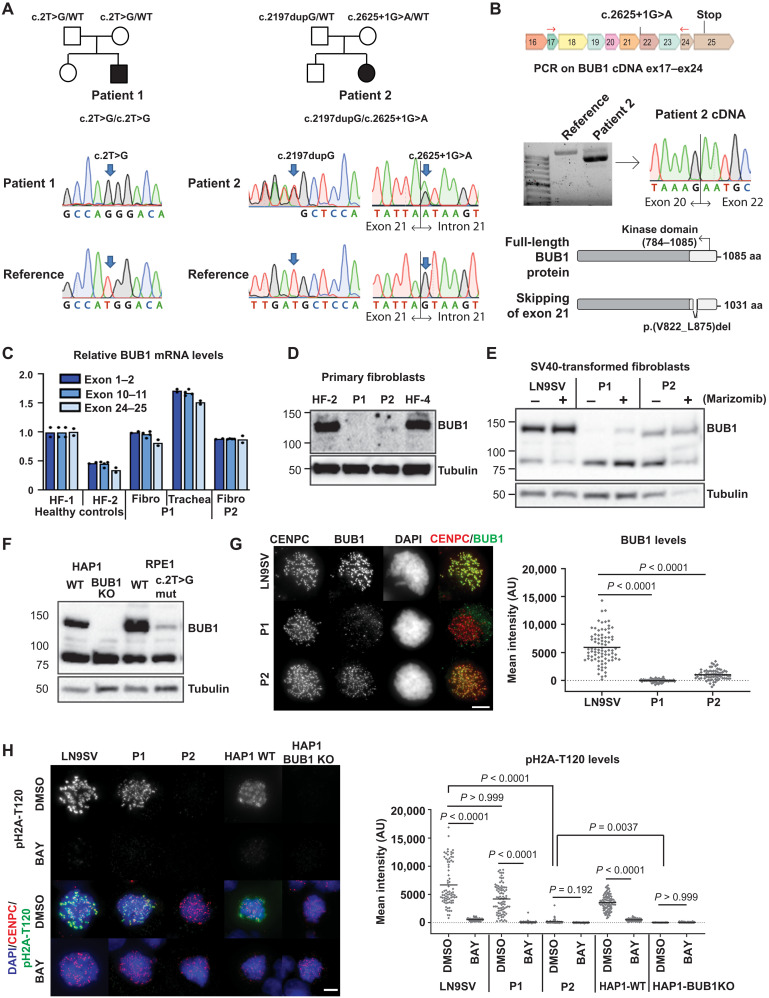
Fig. 1. Biallelic germline mutations in BUB1 cause reduced protein levels.
(A) Family pedigrees (based on WES) and Sanger confirmation from primary fibroblasts. Nomenclature NM_004336.5. (B) P2 primary fibroblast cDNA was PCR-amplified using indicated primers (red arrows) and analyzed on agarose gel and by Sanger sequencing. The transcript lacks exon 21. Predicted effect on BUB1 protein is shown. (C) BUB1 mRNA was assessed in indicated cells using three different primer pairs, each in three technical replicates. (D and E) Western blot of primary fibroblasts (D) (using A300-386A to detect BUB1) and SV40-transformed fibroblasts (E) (using ab9000). CDC6 is a control for marizomib-induced protein stabilization (6 hours, 500 nM). (F) Western blot of engineered RPE1-BUB1 mutant cells and complete BUB1 KO HAP1 cells using ab9000. (G) Colchicine-arrested cells (10 μM, 2 hours) were stained for CENPC, BUB1 (using A300-386A), and DNA. Graph depicts mean BUB1 intensity within CENPC-defined regions. n = 83, 73, and 72 cells from three independent experiments. (H) Colchicine-arrested cells, treated with BUB1 kinase inhibitor BAY1816032 (4 μM, 1 hour) or dimethyl sulfoxide (DMSO), were stained, and mean pH2A-T120 intensity was assessed within DAPI-defined regions. Approximately 90 cells from three independent experiments were analyzed per condition. Each dot represents one cell; black lines indicate the means. Scale bars, 5 μm. Nonparametric Kruskal-Wallis test (Dunn’s multiple comparison test) used for statistical analysis. AU, arbitrary units; aa, amino acid.