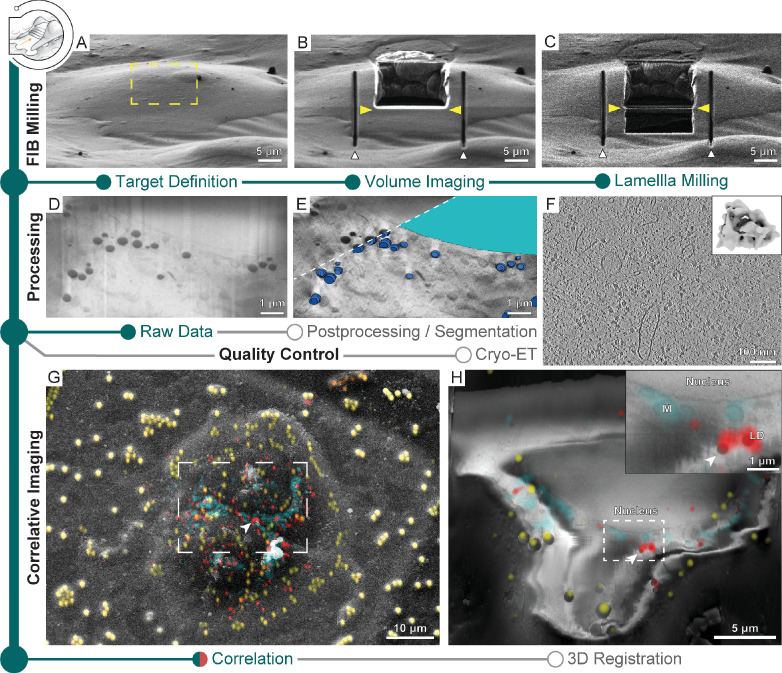
Figure 4. Multi-modal 3D cryogenic imaging by focused ion beam-scanning electron microscopy (FIB-SEM) volume imaging and cryo-electron tomography (cryo-ET).
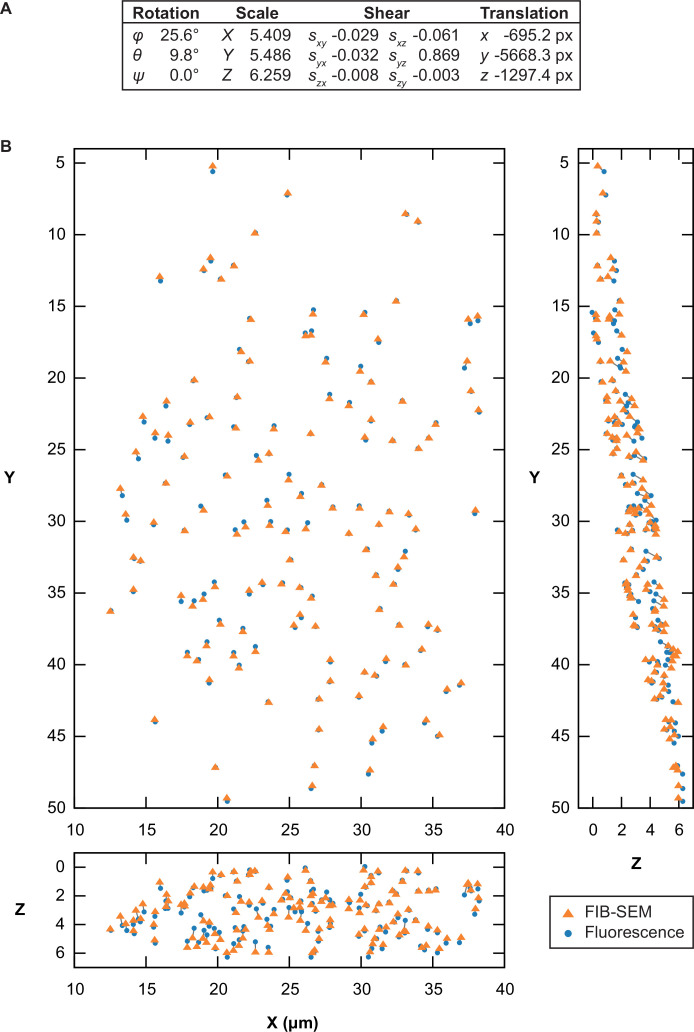
Green modules represent tasks performed with SerialFIB; orange, with 3D Correlation Toolbox (3DCT). Gray modules represent tasks performed externally. (A) Focused ion beam (FIB) image of a Sum159 breast cancer cell prior to milling. Yellow rectangle indicates the volume for FIB-SEM volume data acquisition. (B) FIB image of the cell after FIB-SEM volume imaging. Yellow arrowheads indicate the defined position for lamella preparation. White arrowheads indicate micro-expansion joints prepared after volume imaging. (C) FIB image of the final lamella. (D) Representative slice of the raw FIB-SEM volume data and (E) postprocessed data, overlaid with manual segmentations of lipid droplets (dark blue) and the nucleus (cyan). (F) Slice through a tomogram acquired from lamella in (C). Inset shows the ribosome subtomogram average from data collected on lamellae prepared by this workflow (two tomograms; 3380 subtomograms; 24 Å resolution). (G) SEM view of two HeLa cells overlaid with a fluorescence light microscopy (FLM) maximum intensity projection of lipid droplet (red), mitochondria (cyan), and fiducial bead (yellow) fluorescence signals, determined by correlation in 3D Correlation Toolbox (3DCT). (H) Representative slice through the postprocessed FIB-SEM volume overlaid with a 200 nm virtual slice through the affine-transformed 3D-registered fluorescence volumes. White arrowhead points to a pair of lipid droplets, which are displayed at higher magnification in the inset.