Figure 1 .
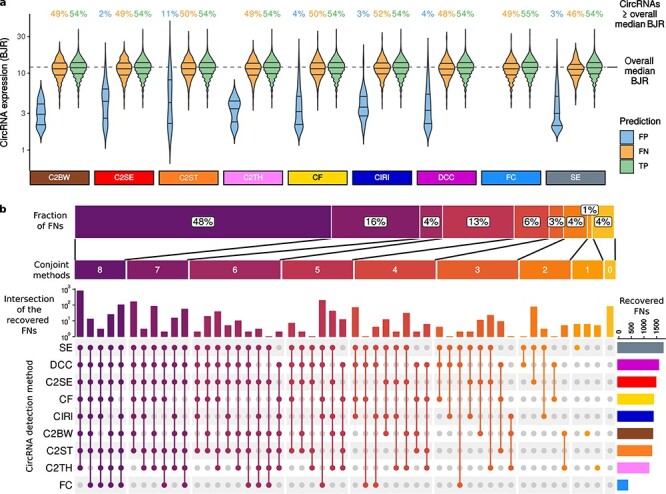
Commonly used circRNA detection methods may overlook some highly expressed circRNAs. A, The expression level of predicted circRNAs. BJR: back-splice junction read counts; C2BW: CIRCexplorer2 on BWA; C2SE: CIRCexplorer2 on Segemehl; C2ST: CIRCexplorer2 on STAR; C2TH: CIRCexplorer2 on TopHat-Fusion; CF: circRNA_finder; CIRI: CIRI2; FC: Findcirc; SE: Segemehl; FP: false-positive; FN: false-negative; TP: true-positive. B, Number of methods detecting circRNAs not detected by other methods and the number of the FNs detected. Colour refers to the number of methods conjointly detecting circRNAs missed by other tools. The vertical bars show the number of FN circRNAs detected by the methods indicated in the coloured dots below the bars. The bars denote disjoint circRNA sets. Grey dots indicate the methods failing to detect the circRNAs considered in the bar chart on the top. The horizontal bars on the right represent the overall number of FN circRNAs detected by the methods. The horizontal bar shows the percentage of detected FNs by grouping method combinations according to the number of combined methods.
