Figure 6 .
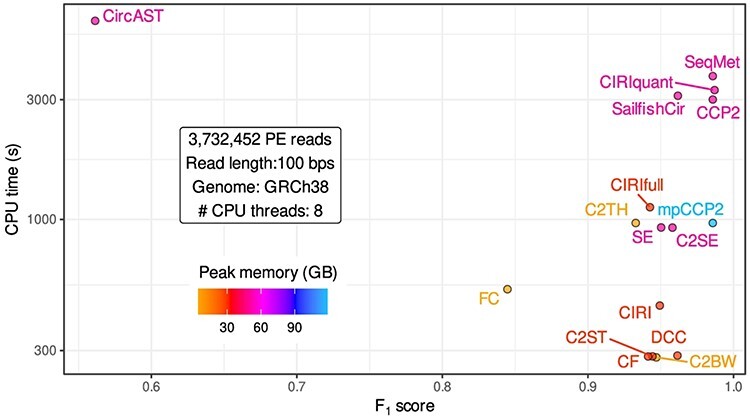
Computation time and memory requirements of circRNA detection methods. F1-scores of 14 circRNA detection pipelines compared to their computing time in seconds (s) and peak random access memory (in GigaBytes) required to process ~3.7 million paired-end (PE) reads of 100 bp length, mapped to the human genome (GRCh38) and using eight CPU threads for each tool. Plus, one hypothetical pipeline running sequentially the seven methods used in CirComPara2 (SeqMet) and CirComPara2 set with maximal parallelization settings (mpCCP2) allowing to run up to seven tasks in parallel, for peak usage of 56 CPU threads in total.
