Table 2.
A comparison of prediction accuracies (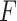 -values) by sequence length for each method
-values) by sequence length for each method
| Length | Short (12–150 nt) | Medium (151–500 nt) | Long (501–4381 nt) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PKF | PK | CB | PKF | PK | CB | PKF | PK | CB | |
| IPknot | |||||||||
| (LinearPartition-C) | 0.681 | 0.552 | 0.258 | 0.492 | 0.482 | 0.128 | 0.433 | 0.428 | 0.061 |
| (LinearPartition-V) | 0.669 | 0.499 | 0.143 | 0.478 | 0.461 | 0.091 | 0.380 | 0.370 | 0.038 |
| (CONTRAfold) | 0.678 | 0.550 | 0.259 | 0.495 | 0.505 | 0.154 | 0.426 | 0.413 | 0.066 |
| (ViennaRNA) | 0.669 | 0.500 | 0.144 | 0.480 | 0.461 | 0.091 | 0.212 | 0.317 | 0.041 |
| ThreshKnot | |||||||||
| (LinearPartition-C) | 0.681 | 0.501 | 0.027 | 0.493 | 0.475 | 0.019 | 0.439 | 0.431 | 0.008 |
| (LinearPartition-V) | 0.669 | 0.484 | 0.033 | 0.481 | 0.456 | 0.026 | 0.383 | 0.372 | 0.014 |
| Knotty | 0.641 | 0.550 | 0.315 | — | — | — | — | — | — |
| SPOT-RNA | 0.658 | 0.621 | 0.322 | 0.462 | 0.479 | 0.127 | — | — | — |
| CONTRAfold | 0.682 | 0.519 | 0.000 | 0.500 | 0.497 | 0.000 | 0.425 | 0.415 | 0.000 |
| RNAfold | 0.668 | 0.472 | 0.000 | 0.474 | 0.442 | 0.000 | 0.361 | 0.347 | 0.000 |
PKF, 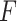 -value for pseudoknot-free sequences; PK,
-value for pseudoknot-free sequences; PK, 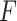 -value for pseudoknotted sequences; CB,
-value for pseudoknotted sequences; CB, 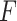 -value of crossing base pairs.
-value of crossing base pairs.
