FIG 1.
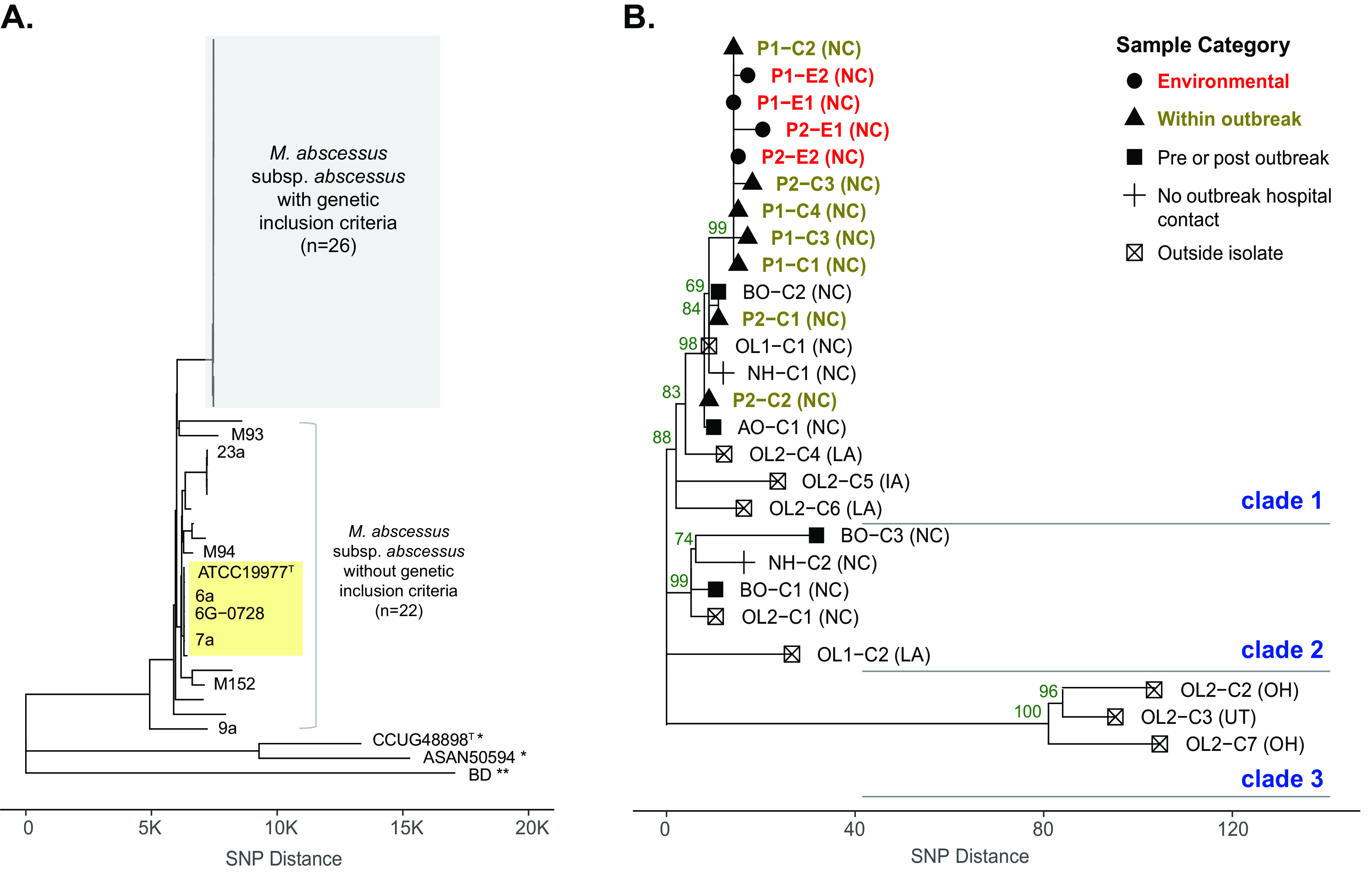
Phylogenomic analysis of study cohort including outbreak and control M. abscessus subsp. abscessus isolates. (A) Genome-wide SNP data from study isolates with genetic inclusion criteria (n = 26) and isolates without genetic inclusion criteria (n = 22) were analyzed using the neighbor-joining (NJ) method. The x axis represents SNP distance. Outgroup isolates of M. abscessus subsp. massiliense (*) and M. abscessus subsp. bolletii (**) were included. The clade with the previously described “dominant circulating clone,” including the M. abscessus subsp. abscessus type strain ATCC 19977T, is labeled in yellow. (B) Study isolates with genetic inclusion criteria were analyzed in detail. Sample categories are designated shapes, and color-coded labels are indicated for environmental (red) and within outbreak (brown) isolates. Bootstrap support values from 100 replicate searches are shown on selected nodes (values of >50), and three genetic clades are labeled. Suspected U.S. state of acquisition is given in parentheses. Abbreviations: AO, after outbreak; BO, before outbreak; C, clinical; E, environmental; NH, neighboring hospital; OL, outside laboratory; P, phase of outbreak.
