Figure 5. Highly efficient genome editing in cardiovascular ECs in adult mice.
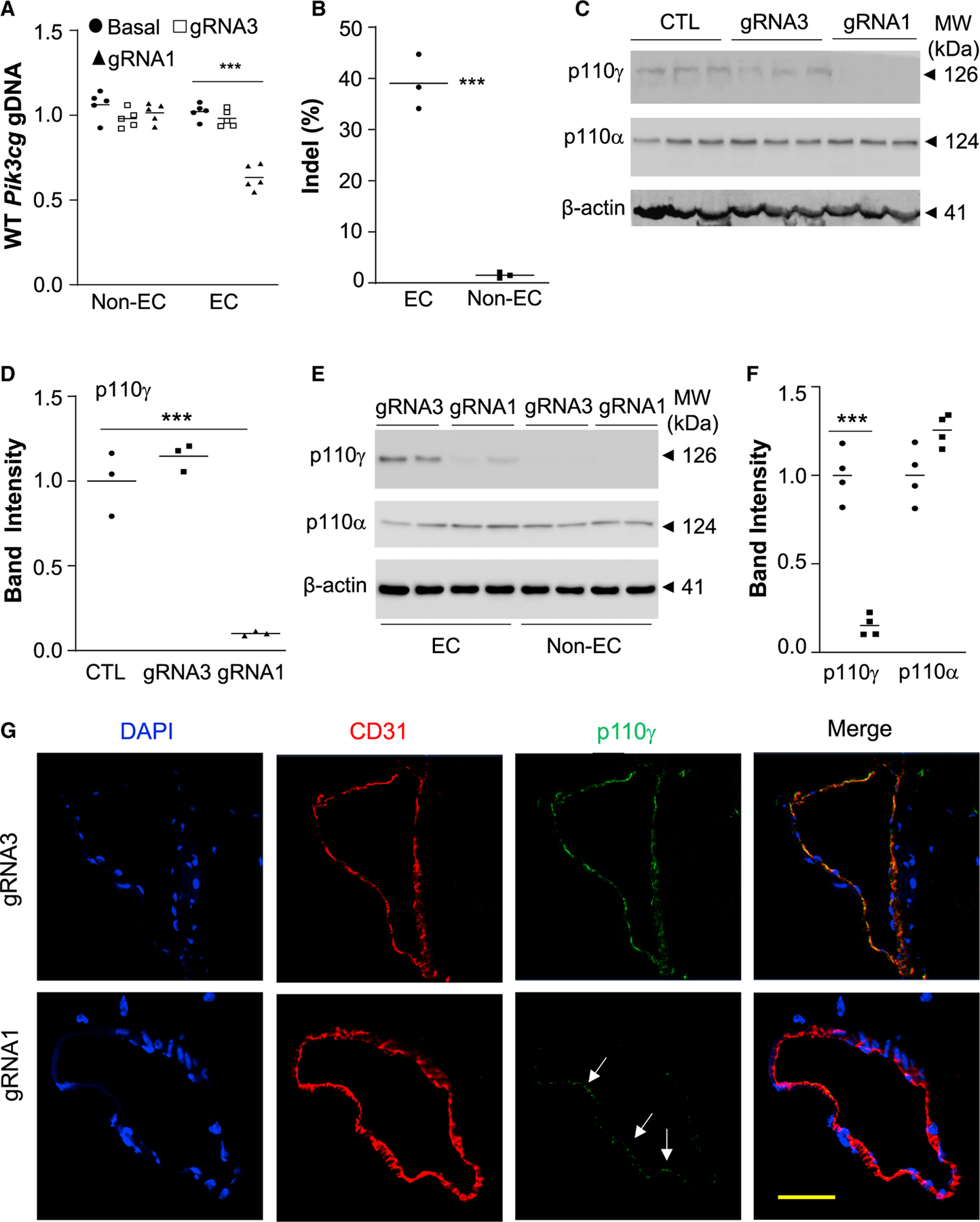
(A) QPCR analysis demonstrating robust genome editing in cardiovascular ECs but not in non-ECs of Pik3cg gRNA1 nanoparticle-treated mice. At 7 days post-nanoparticle delivery, hearts were collected for EC isolation with anti-CD31 magnetic beads. ***p < 0.001, one-way ANOVA with Bonferroni post hoc analysis (n = 5).
(B) Sanger sequencing decomposition analysis showing indels exclusively in cardiovascular ECs at a rate of 40% (n = 3).
(C, D) Western blotting demonstrating greater than 80% decrease of p110γPI3K protein expression in hearts of CRISPRCDH5/gRNA1 nanoparticle-transduced mice compared with control (CTL) mice without plasmid DNA transduction. Quantification was carried out with Image J and normalized to loading control β-actin (n = 3).
(E, F) Diminished expression of p110γPI3K in cardiovascular ECs of Pik3cg gRNA1 nanoparticle-treated mice (n = 4).
(G) Representative immunofluorescent micrographs showing diminished p110γPI3K expression in cardiovascular ECs of gRNA1 nanoparticle-treated mice. Cryosections of mouse hearts were immunostained with anti-p110γ and anti-CD31 antibodies. Arrows point to ECs with less efficient knockdown of p110γPI3K. Scale bar, 50 μm. ***p < 0.001, one-way ANOVA with Bonferroni post hoc analysis (A, D), Student’s t test (B, F).
