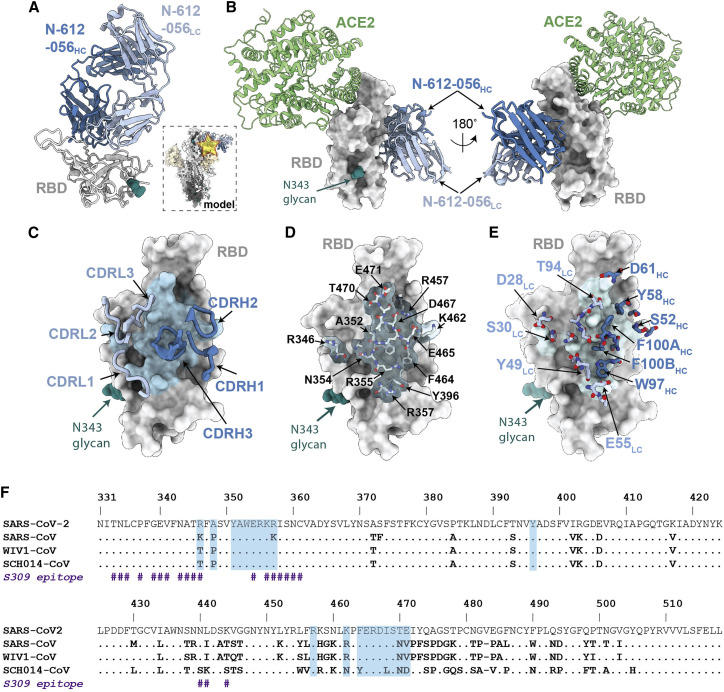
Figure 4.
X-ray crystal structure of SARS-CoV-2 RBD in complex with the N-612-056 Fab
(A) 2.9 Å X-ray crystal structure for the N-612-056 Fab-RBD complex. Inset: overlay of the N-612-056-RBD crystal structure on an S trimer with up RBD conformation (PDB: 6VYB).
(B) Composite model of N-612-056-RBD (blue ribbon and gray surface, respectively) overlaid with soluble ACE2 (green; PDB: 6M0J). The model was generated by aligning RBDs on 191 matched Cα atoms.
(C) N-612-056 CDR loops (blue) mapped on the RBD surface (gray). The N-612-056 epitope is shown as a light blue surface.
(D) Surface and stick representations of N-612-056 epitope.
(E) N-612-056 paratope residues mapped on the RBD surface with epitope residues shown in light blue.
(F) Sequence alignment of SARS-CoV-2, SARS-CoV, WIV1-CoV, and SCH014-CoV. N-612-056 epitope residues are shaded blue. S309 epitope residues are also shown (indicated with the hash [#] symbol).