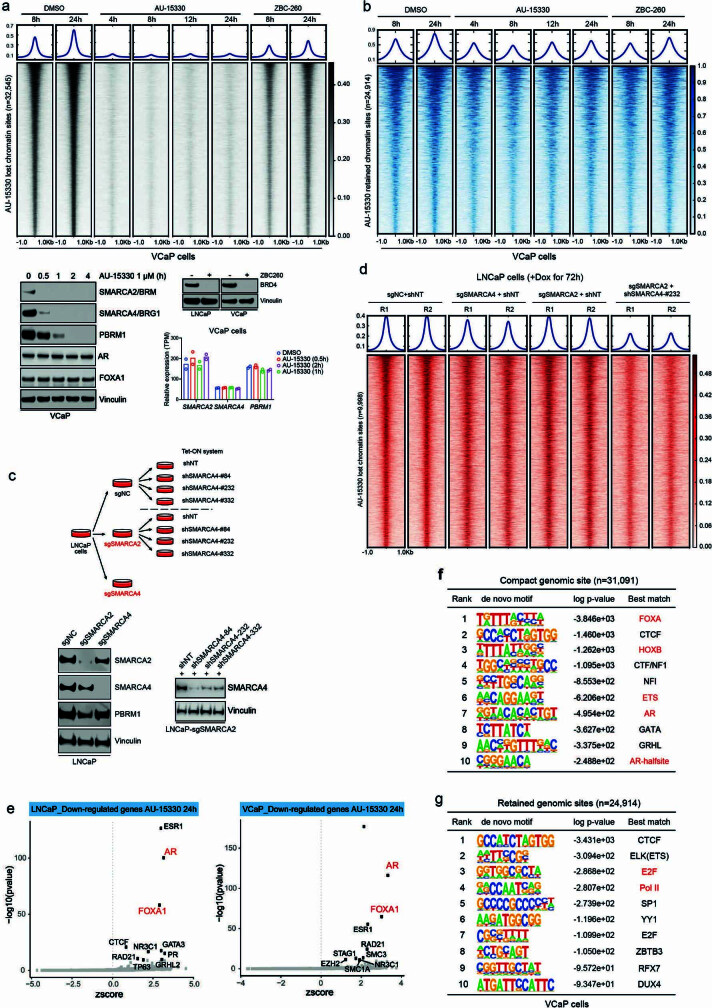
Extended Data Fig. 3. SWI/SNF ATPases, SMARCA2 and SMARCA4, mediate chromatin accessibility at numerous sites across the genome in PCa cells.
(a, b) ATAC-seq read-density heatmaps from VCaP cells treated with DMSO (solvent control), AU-15330, or ZBC-260 (a BRD4 degrader) for indicated durations at genomic sites that are compacted (a) or remain unaltered (b) upon AU-15330 treatment. Immunoblots show loss of target proteins upon treatment of cancer cells with AU-15330 (1 μM) for increasing durations or ZBC-260 (10 nM) for 4h. Vinculin is the loading control probed on all immunoblots. This experiment was repeated independently twice. Barplot shows the changes in mRNA expression (RNA-seq) of AU-15330 (1 μM) target genes in VCaP cells treated for noted durations. (c) Schematic outlining the CRISPR/Cas9 and shRNA-based generation of LNCaP cells with either independent or simultaneous inactivation of SWI/SNF ATPases, SMARCA2 and SMARCA4. Immunoblots showing the decrease in target expression in the genetic models shown above. Vinculin is the loading control probed on a representative immunoblot. This experiment was repeated independently twice. (d) ATAC-seq read-density heatmaps from genetically engineered LNCaP cells with SMARCA2 and/or SMARCA4 functional inactivation at AU-15330-compacted genomic sites. (e) Binding analysis for the regulation of transcription (BART) prediction of specific transcription factors mediating the observed transcriptional changes upon AU-15330 treatment in LNCaP or VCaP cells. The top 10 significant and strong (z-score) mediators of transcriptional responses are labeled (BART, Wilcoxon rank-sum test). (f) Top ten de novo motifs (ranked by p-value) enriched within AU-15330-compacted genomic sites (HOMER, hypergeometric test) in VCaP cells. (g) De novo motif analysis with top 10 motifs (ranked by p-value) enriched within genomic sites that retain chromatin accessibility upon AU-15330 treatment in VCaP cells (HOMER hypergeometric test).