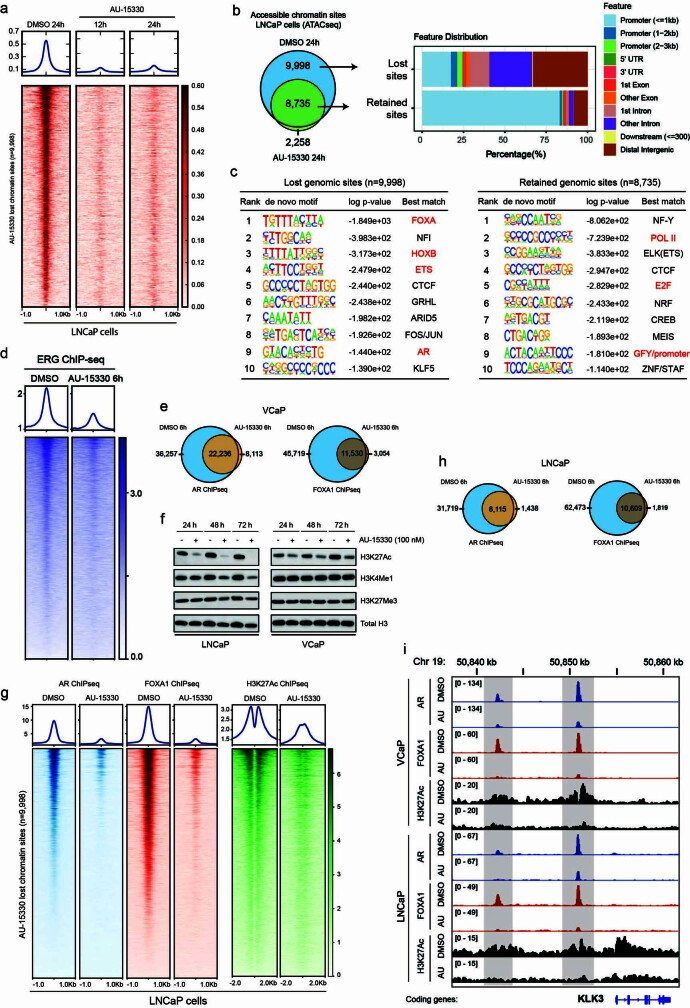
Extended Data Fig. 4. SWI/SNF inhibition condenses chromatin at enhancer sites bound by oncogenic transcription factors AR and FOXA1 in PCa cells.
(a) ATAC-seq read-density heatmaps from LNCaP cells treated with DMSO or AU-15330 for indicated durations at all genomic sites that lose physical accessibility upon AU-15330 treatment. (b) Genome-wide changes in chromatin accessibility upon AU-15330 treatment for 12h in LNCaP cells, along with genomic annotation of sites that are lost or retained in the AU-15330-treated cells. (c) De novo motif analysis with top 10 motifs (ranked by p-value) enriched within AU-15330-compacted or unaltered genomic sites in LNCaP cells (HOMER, hypergeometric test). (d) ChIP-seq read-density heatmaps for ERG at the AU-15330-compacted genomic sites in VCaP cells after treatment with DMSO or AU-15330 (1 μM) for indicated times and stimulation with R1881 (1 nM, 3h). (e) Genome-wide changes in AR and FOXA1 ChIP-seq peaks upon AU-15330 treatment (1 μM, 6h) in VCaP cells stimulated with R1881, a synthetic androgen (1 nM, 3h). (f) Immunoblots showing the changes in indicated chemical histone marks upon treatment with AU-15330. Vinculin is the loading control probed on a representative immunoblot. This experiment was repeated independently twice. (g) ChIP-seq read-density heatmaps for AR, FOXA1, and H3K27Ac at the compacted genomic sites in LNCaP cells after indicated durations of treatment with AU-15330 (1 μM). (h) Genome-wide changes in AR and FOXA1 ChIP-seq peaks upon AU-15330 treatment (1 μM, 6h) in LNCaP cells stimulated with R1881 (1 nM, 3h). (i) ChIP-seq tracks for AR, FOXA1, and H3K27Ac within the KLK2/3 gene locus in R1881-stimulated VCaP and LNCaP cells with or without AU-15330 (AU).