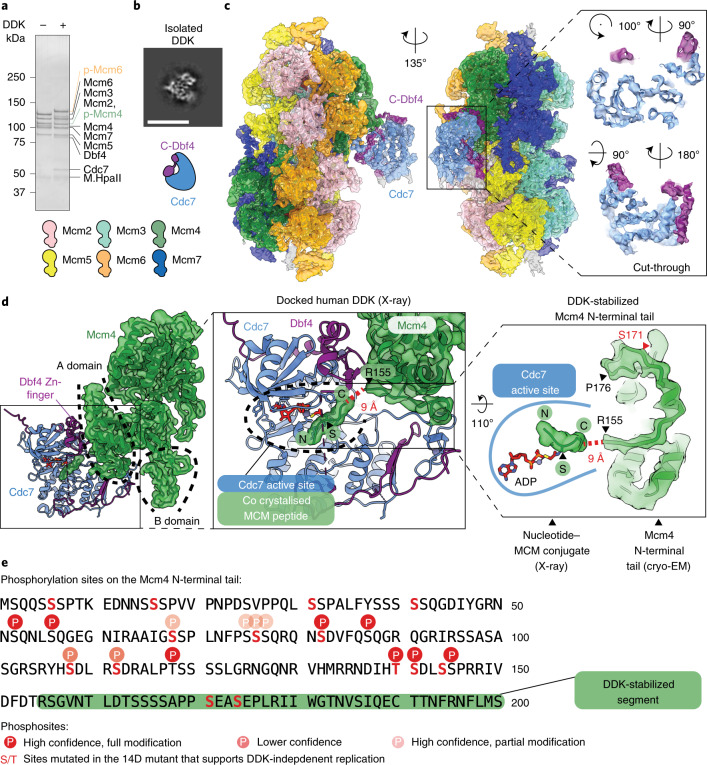
Fig. 2. Molecular basis for kinase recognition and phosphorylation of the MCM DH substrate.
a, SDS–PAGE gel of unmodified and DDK-phosphorylated MCM DHs, tethered to DNA beads. The phosphorylation-dependent shifts of Mcm4 and Mcm6 are highlighted in green and orange, respectively. Notably, after a low-salt-wash step, DDK remains bound to the MCM. Representative of at least n = 3 independent experiments. b, 2D class average of the isolated DDK shows subnanometer-resolution features, indicating that DDK is a suitable cryo-EM target. Scale bar, 10 nm. c, 3.3-Å-resolution structure of the MCM DH–DDK complex, showing the catalytic core of DDK (Cdc7 bound to C-terminal Dbf4), engaged to the Mcm4 subunit of one MCM ring in the DH. Two cut-through views of the kinase core are shown to highlight how the atomic model matches cryo-EM density. d, DDK docks onto the Mcm4 A domain via the Dbf4 zinc finger C domain, and onto the Mcm4 B domain via the Dbf4 M domain. Middle: active site of the human DDK crystal structure, which was cocrystallized with an MCM substrate peptide in the active site of Cdc7. The first resolved N-terminal residue (R155) of Mcm4 in the cryo-EM map neatly aligns with the C-terminal end of the MCM peptide. The Mcm4 N-terminal tail in our structure is therefore suitably poised for phosphorylation by the Cdc7 active site. An N-terminal Mcm4 segment (P155 to R176), which is invisible in the absence of DDK, is partially stabilized in the DH–DDK complex. e, One known DDK phosphosite in Mcm4 (S171) maps within the DDK-stabilized N-terminal segment visible in our structure. Five additional known phosphosites and others detected by mass spectrometry map upstream of the modeled N-terminal region of Mcm4. This agrees with the notion that active site access requires extended structural flexibility of the phosphorylation substrate. Sites reported to be important for recruitment of the firing factor Sld3 are highlighted. The uncropped gel image for a is available as Source data with the paper online.