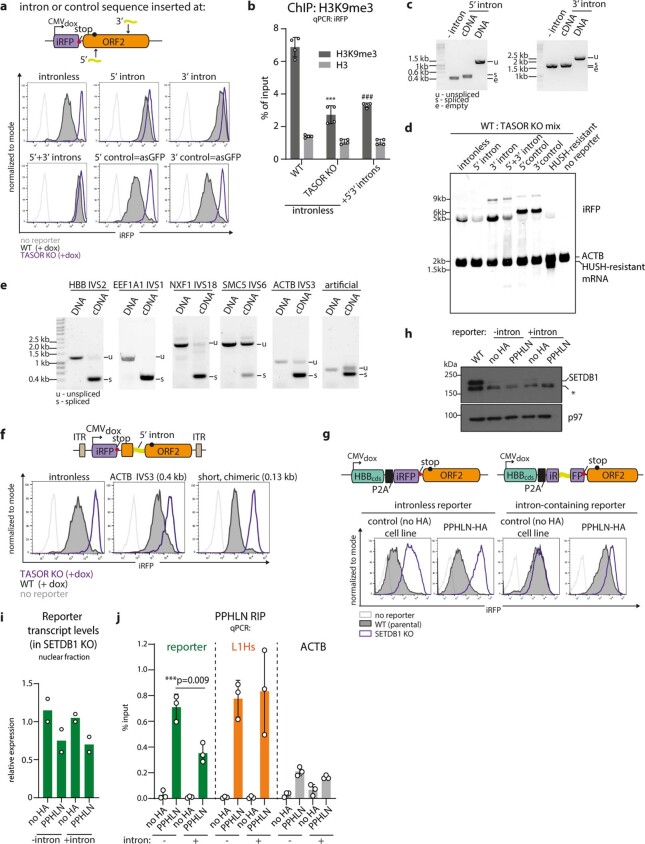
Extended Data Fig. 7. Intron insertion reduces HUSH-mediated repression and Periphilin binding to reporter transcripts.
a, Representative flow cytometry histograms of expression from reporters in Fig. 3c in WT and TASOR KO HeLa cells. The 5’ and 3’ control (asGFP) is the antisense GFP ‘stuffer‘ sequence b, ChIP-qPCR quantifying H3K9me3 and total H3 levels at intronless or reporter with introns (HBB IVS2) inserted at 5’ and 3’ of ORF2 (from Fig. 3c and Extended Data Fig. 7a). n = 4 independent experiments ± SD; ***p = 0.0003 and ###p = 0.0006 vs –intron WT, ratio paired two-tailed t-test. c, Gel images confirming splicing of introns at 5’ and 3’ of ORF2 from iRFP-ORF2 reporter transcripts (from Fig. 3c and Extended Data Fig. 7a) by PCR. d, Northern blot analysis of mRNAs produced from reporters with intron or control sequence inserted 5‘and 3’ of ORF2 (~5kb), or HUSH-resistant reporter without ORF2 (~1.5kb). RNA was isolated from the mix of WT and TASOR KO cells. e, PCR analysis of splicing of different introns 5’ of ORF2 from iRFP-ORF2 reporter transcripts (from Fig. 3d and Extended Data Fig. 7f). f, Representative flow cytometry histograms of expression from iRFP-ORF2 reporter with introns from ACTB (0.4kb) or a short, chimeric intron (0.13kb) cloned 5’ of ORF2 in WT and TASOR KO HeLa cells. Experiment repeated independently with similar results; quantification of n = 3-4 biological replicates in Fig. 3f. g, Schematic of intronless and intron-containing reporter constructs for periphilin RIP-qPCR (upper schematic). Reporters were integrated into WT 293T or periphilin-HA 293Ts - resulting in four independent cell lines. SETDB1 function was disrupted by CRISPR/Cas9-mediated knockout and mixed, polyclonal KO populations were used for RIP-qPCR. Flow cytometry histograms of expression from reporters in PPHLN-HA and control cell lines 48h after induction with dox (bottom). h, Validation of SETDB1 depletion by CRISPR/Cas9 in PPHLN1 HA-KI cells by western blot. β-actin as loading control. * marks non-specific band. i, Relative levels of transcripts from reporters for RIP-qPCR (in SETDB1 KO) in nuclear fraction normalized to ACTB; n = 2 technical replicates. j, RIP-qPCR showing decreased association of periphilin with RNA from intron-containing reporter. L1Hs and ACTB RNA are a positive and negative control, respectively. Data are mean ± SD; n = 3 independent experiments; and normalized to input.***p = 0.0009 vs -intron, one-way ANOVA post-hoc pairwise comparison with Bonferroni correction.