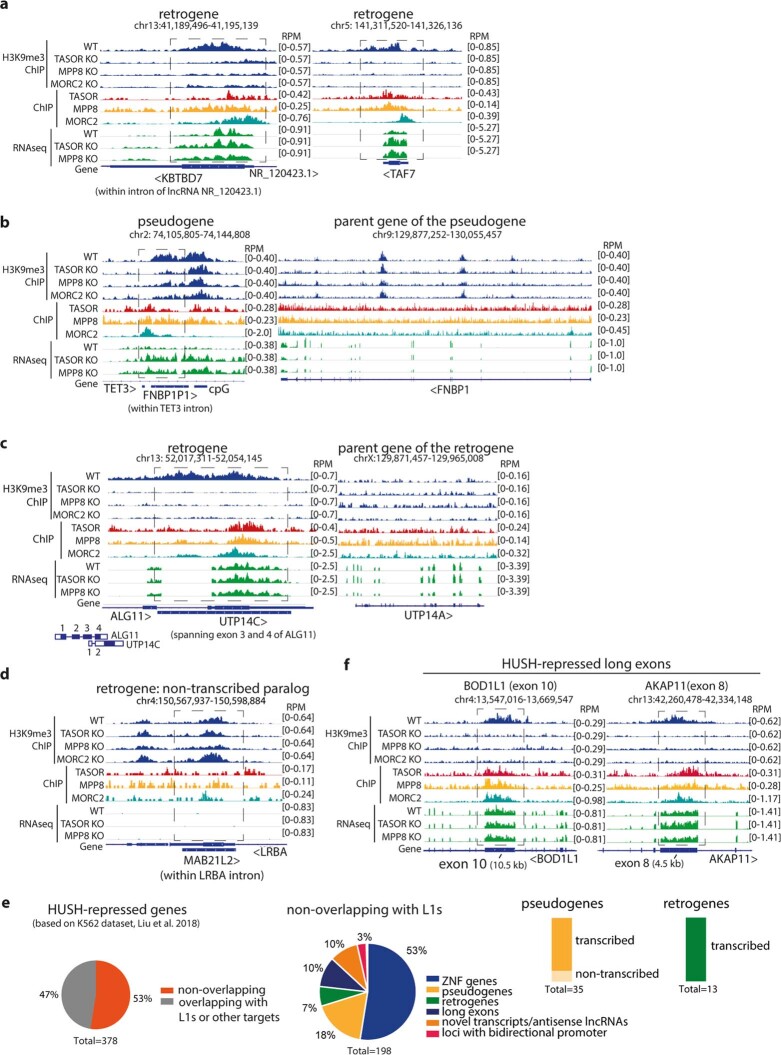
Extended Data Fig. 9. Transcribed processed pseudogenes and protein coding retrogenes, but not their parent genes, are bound and silenced by HUSH.
Genome browser tracks showing HUSH-dependent H3K9me3, HUSH/MORC2-occupancy and RNA-seq in WT and HUSH KO K562 cells at: a, additional, representative loci of retrogenes; b, FNBP1P1 pseudogene (left) and its parent gene FNBP1 (right); c, UTP14C retrogene (left) and its parent gene UTP14A (right) d, at the locus of MAB21L2, a non-transcribed paralog of HUSH-repressed MAB21L1 retrogene, Data from 2. e, HUSH-repressed genes obtained from the dataset in ref. 2: 378 genes were obtained when there was at least 30% reduction of H3K9me3 signal in all 3 knockout cell lines: TASOR KO, MPP8 KO and MORC2 KO (log2 FC H3K9me3 TASOR KO/WT ≤ -0.5; FDR significance ≤ 0.05; determined after a comparative assessment of counts between conditions (n = 2) using negative binomial generalized linear models as implemented in edgeR and corrected for multiple comparisons using FDR method). 104 of them were ZNF genes (including 8 ZNF pseudogenes) and the rest were inspected in IGV to determine the most probable reason for HUSH-repression e.g. overlap with L1 elements or other HUSH targets, pseudogene, retrogene, genes with signal over long exons, novel transcripts or antisense lncRNAs or loci with bidirectional promoter. The 11 resulting genes remained unannotated, either excluded because of low, background H3K9me3 over region or mapping artifacts or the reason for repression was unclear. Fraction of pseudogenes and retrogenes transcribed (average RNA-seq signal of all samples above > 0.1 RPKM or within transcriptionally active gene) (right) f, Genome browser tracks showing HUSH-dependent H3K9me3, HUSH/MORC2-occupancy and RNA-seq in WT and HUSH KO K562 cells at representative long exons. Data from ref. 2.