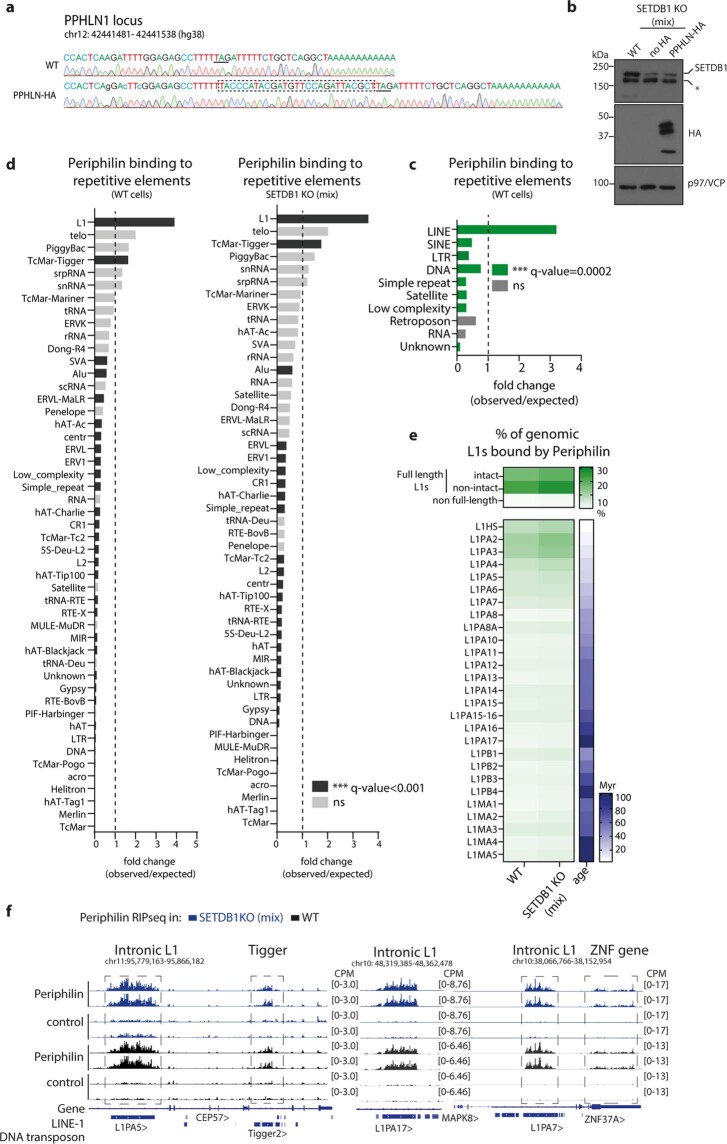
Extended Data Fig. 5. Periphilin specifically binds transcripts from evolutionary young, full-length L1 elements in WT and SETDB1-depleted cells.
a, Sequencing tracks showing insertion of sequence of HA-tag (marked as dashed box) into PPHLN1 locus. Underlined is the stop codon. Nucleotide substitutions to make modified locus sgRNA-resistant are marked as small letters. b, Western blot showing SETDB1 depletion in SETDB1 KO (mix) Periphilin-HA HEK293Ts and control HEK293Ts. p97/VCP as a loading. c,d, Enrichment of periphilin RIP-seq peaks at different repetitive elements in c, WT cells and d, WT (left) and SETDB1 KO (mix) (right). Significant enrichment is defined as a fold change score greater than one with Benjamini–Hochberg empirical adjusted one-sided p-value calculated using simulations and genomic association testing47, ***q < 0.001 (for exact p-values see source data). e, Fraction of full length, non-full length L1s and L1s from different families overlapping with periphilin RIPseq peaks. Full length L1s definitions are based on L1Base48,49. Blue heatmap indicates age of L1 families predicted from the phylogenetic analysis50. Periphilin-bound L1Hs may be underestimated in comparison to L1PA2-L1PA3 due to lower mappability of L1Hs as this is the least sequence-divergent L1 family. f, Genome browser tracks showing periphilin RIP signal over intronic L1s, Tigger DNA transposon and 3’UTR of ZNF37A