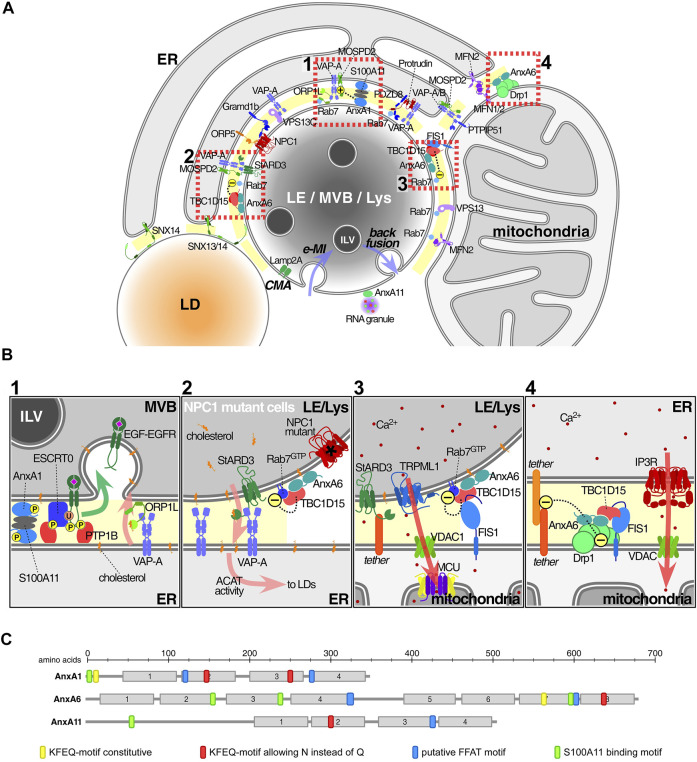
FIGURE 1.
Annexins and associated protein complexes at membrane contact sites. (A) Endolysosomes (LE/MVBs/Lys) establish multiple membrane contact sites (MCS) with a variety of other organelles, in particular the ER and mitochondria, but also peroxisomes and lipid droplets. Here we only illustrate a subset of contacts and contact site proteins. Lipid transfer, including cholesterol as well as Ca2+ mobilization and signalling are probably the most important MCS-associated functions. Annexins have been found in MCS connecting the ER with either LE or mitochondria (MAMs), as well as in LE-mitochondria contacts. Other proteins or protein complexes serve as tethers to ensure the formation of MCS, including several sorting nexins (Henne et al., 2015; Dong et al., 2016; Saric et al., 2021), VAP proteins, PTPIP51, PDZD8 or protrudin (Shirane et al., 2020; Neefjes and Cabukusta, 2021). Furthermore, ORP5/8 and NPC1/Gramd1 also translocate lipids including PS and cholesterol, across MCS between the ER and mitochondria and/or LE respectively (Galmes et al., 2016; Hoglinger et al., 2019). At the bottom of this endolysosome, we contemplate the possible recruitment of cytosolic annexins into ILV (see text for details) via two autophagic pathways. While chaperone-mediated and Lamp2a-dependent autophagy (CMA) may carry annexins into the lumen of MVBs or the outer membrane of ILVs, endosomal microautophagy (e-MI) may board annexins inside ILV (both routes via KFERQ-motif) (Tekirdag and Cuervo, 2018). In any case, whatever the topology, these proteins have to escape from lysosomal degradation (Meneses-Salas et al., 2020b). Then, ILV might undergo back fusion/retrofusion (Eden and Futter, 2021), delivering annexins and/or other cargo out of the MVB into the cytosol in the vicinity of MCS, where a suitable local Ca2+ and lipid microenvironment could then favour retention in MCS. Alternatively, MVB diversion to exocytosis could generate exosomes. This could be the destination of AnxA11, highly enriched in exosomes. Otherwise, cytosolic AnxA11 could be confined to PI(3,5)P2 at the endolysosomal cytosolic membrane to tether RNA granules (Liao et al., 2019). (B) 1: AnxA1, together with S100-A11 as a tetrameric complex, tethers MCS to mediate cholesterol transport from ER to LE/Lys via interaction of VAP-A with ORP1L (Eden et al., 2016). In this scenario, AnxA1 overexpression increases MCS between MVB and ER (Wong L. H. et al., 2018). 2: AnxA6 overexpression decreased MCS numbers between ER and LE/Lys, whereas AnxA6 depletion in NPC1 mutant cells stimulated MCS formation. The underlying mechanism involves the recruitment of AnxA6 and TBC1D15 to Rab7-positive organelles and was associated with increased LE motility and LE-cholesterol release into the ER, through StARD3-VAP-A in MCS (Meneses-Salas et al., 2020b). 3: TBC1D15 and Rab7 in complex with FIS1 between LE/Lys and mitochondria affects the fission of mitochondria (Wong Y. C. et al., 2018). 4: AnxA6 interacts with Drp1 and FIS1 between ER and mitochondria to modulate Ca2+ dynamics and mitochondrial fission (Chlystun et al., 2013). In all settings shown in Insets 2-4 the presence of AnxA6 seems to cause MCS untethering. Arrows indicate the following: translocation of EGFR-EGF into ILV (green, inset 1), cholesterol flux (pink, insets 1-2), Ca2+ flux (red, insets 3-4). (C) Schematic representation of the domain structure of the three annexins found in MCS: Motifs that may be involved in the recruitment annexins to MCS are indicated and include the homology to FFAT motifs (blue), S100-binding sites (green) (Rety et al., 2000; Chang et al., 2007; Rintala-Dempsey et al., 2008) and KFERQ-motifs (yellow and red) (Cuervo et al., 2000). Abbreviations that do not appear in the text: ACAT, Acyl-CoA:cholesterol acyltransferase; MCU, mitochondrial calcium uniporter; FIS1, mitochondrial fission 1; IP3R, inositol 1,4,5-triphosphate receptor; TRPML1, transient receptor potential mucolipin 1; Vps13, Vacuolar protein sorting-associated protein 13; MFN1/2, mitofusin1/2; Gramd1b, GRAM domain containing 1B; VAP proteins, VAP-A, VAP-B; monomer specific d-peptide 1 (MOSD1, 2 and 3) and PS, phosphatidylserine.