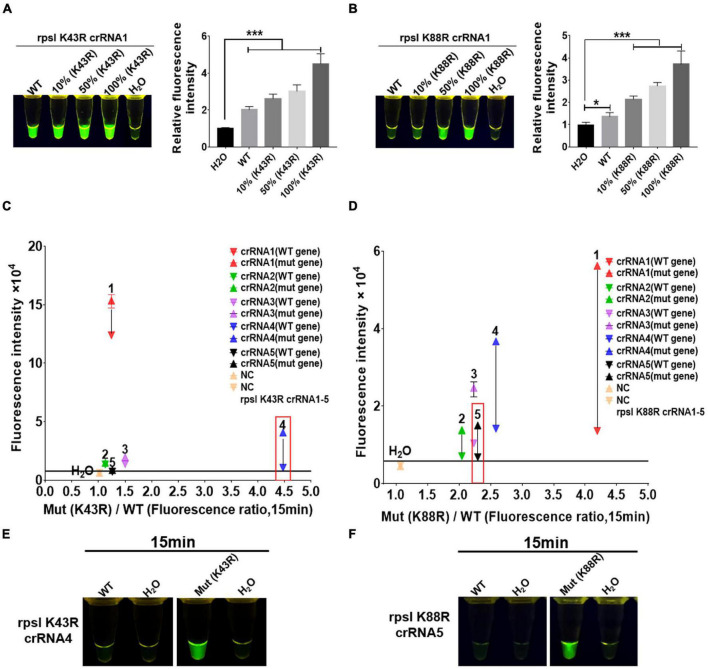
FIGURE 3.
Screening and optimization of the crRNA at rpsl K43R and rpsl K88R streptomycin (STR)-resistant mutation sites. The Cas12a RR system detects the efficiency of the rpsl gene-specific crRNA of STR-resistant M.tb. The fluorescent images and intensities (A,B) at 15 min of the reaction are shown. The rpsl gene fragments of wild-type M. tuberculosis and streptomycin-resistant M. tuberculosis (K43R or K88R) were mixed in different proportions to obtain the detection samples with mutation rates of 100, 50, and 10%, respectively. (C,D) Specificity was determined as the ratio of the on-target DNA-induced fluorescence intensity over the off-target DNA-induced fluorescence intensity. The fluorescence intensity was obtained from crRNA–Cas12a RR reaction for 15 min and measured using a microplate reader. The core crRNA sequences are listed in Supplementary Table 2. (E,F) The Cas12a RR system verifies the specific crRNA of the STR-resistant M.tb rpsl gene fragment and shows the fluorescence image at 15 min of reaction. The concentration of the target gene used in the above-mentioned detection reaction is 1 × 1011 copies. Data are presented as the mean ± SEM from at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.