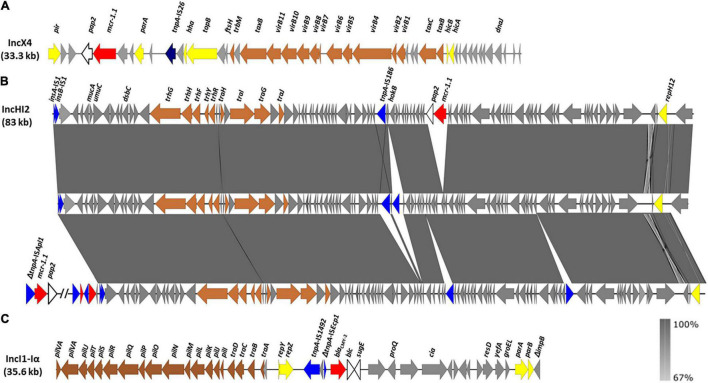
FIGURE 2.
Genetic environment of the mcr-1.1 and blaCMY–2 genes detected in food-borne isolates of the European monophasic clone. (A) Genetic context of mcr-1.1 carried by IncX4 plasmids in the LSP 237/15, LSP 295/15, and LSP 298/15 isolates. (B) Genetic organization of a ca. 83 kb contig carrying the mcr-1.1 gene in the IncHI2 plasmid of LSP 38/19. The alignment of this contig with homologous regions in two related plasmids either lacking mcr-1.1 (pKUSR18; accession number KM396298) or having the gene outside this region (pSE08-00436-1; accession number NZ_CP020493) was created with EasyFig BLASTn. The gray shading between the regions reflects nucleotide sequence similarity ranging from 67 to 100%, according to the scale shown at the lower right part of the figure. (C) Genetic environment of the blaCMY–2 gene in a 35.6 kb contig of the IncI1-I(α) plasmid of LSP 38/19. The open reading frames (ORFs) are represented by arrows pointing to the direction of transcription and having different colors according to their function: red, resistance; yellow, plasmid replication, stability and maintenance; brown, conjugation; blue, DNA mobility, with the tnpA gene of IS26 shown in dark blue; gray, other roles; white, pap2, adjacent to mcr-1.1 in the IncX4 and IncHI2 plasmids, and bcl-sugE, adjacent to blaCMY–2 in the IncI1-I(α) plasmid. The scale is different for (A–C).