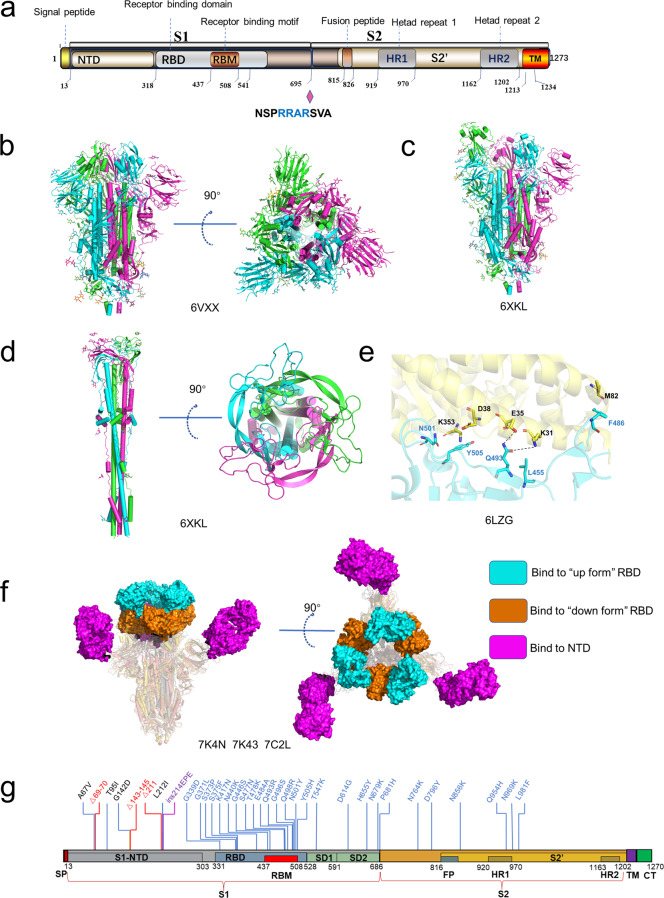
Fig. 3. The structures of S protein.
a Schematic of SARS-CoV-2 S protein domain architecture. The S1 and S2 subunits are indicated, with diamond representing the locations of furin cleavage site. SP signal peptide, RBD receptor-binding domain, RBM receptor-binding motif, SD1 subdomain 1, SD2 subdomain 2, FP fusion peptide, HR1 heptad repeat 1, HR2 heptad repeat 2, TM transmembrane region, CT cytoplasmic tail. b Side and top views of the prefusion structure of the SARS-CoV-2 S protein with all RBD in the “down” conformation (PDB: 6VXX). Protein is shown in cartoon representation, and glycosyls are shown as sticks. c Side view of the prefusion structure of the SARS-CoV-2 S protein with one RBD in the “up” conformation (PDB: 6XKL). d Side and top views of the post-fusion structure of the SARS-CoV-2 S protein (PDB: 6M3W). e Structure of the SARS-CoV-2 RBD complexed with ACE2 (PDB: 6LZG). ACE2 is shown in yellow. The RBD is shown in cyan. Key contacting residues are shown as sticks at the SARS-CoV-2 RBD–ACE2 interfaces. f Side and top views of superimposed three cryo-EM structures of SARS-CoV-2 S in complex with nAbs. S2E12 (represented as a cyan surface) binds to the “up” conformation of SARS-CoV-2 S RBD (PDB: 7K4N); S2M11 (represented as a brown surface) binds to the “down” conformation of SARS-CoV-2 S RBD (PDB: 7K43); 4A8 (represented as a magento surface) binds to the NTD of SARS-CoV-2 S (PDB: 7C2L). g Amino acids mutated in the Omicron variant S protein.