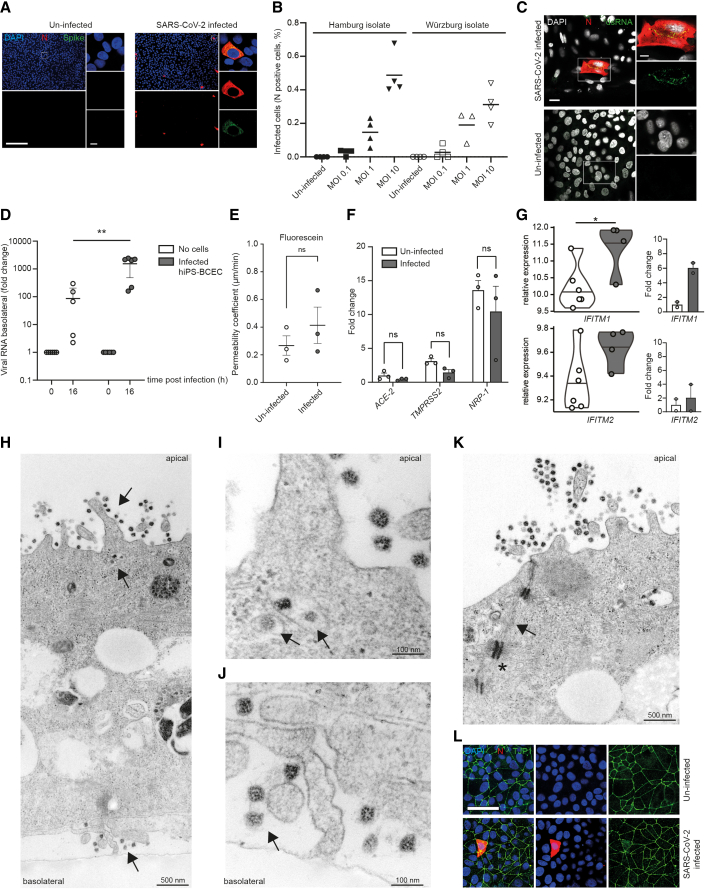
Figure 2.
SARS-CoV-2 infects and replicates in hiPS-BCECs
(A) Representative overview images that were used for subsequent quantification and respective close ups of N and spike protein double staining after infection with SARS-CoV-2 for 24 h (MOI 10) are shown. In the overview images, N protein is oversaturated to enable easy counting of infected cells; the close ups display the subcellular localization of N and spike protein in infected cells. Uninfected cells served as control and did not show any staining with SARS-CoV-2-specific antibodies. SARS-CoV-2 N protein (red), SARS-CoV-2 spike protein (green), counterstained by DAPI (blue). Scale bar, 200 μm; close up, 7.5 μm.
(B) Infected hiPS-BCECs (MOI 0.1, 1, and 10 each for Hamburg and Würzburg isolates) stained for N protein, indicating a dose-dependent rate of infection; n = 2 independent experiments, three or four technical replicates per condition.
(C) Representative immunofluorescence of SARS-CoV-2-infected hiPS-BCECs stained for N protein (red) and double-stranded RNA (dsRNA, green) (MOI 10), counterstained by DAPI (white). Uninfected hiPS-BCECs served as control. Scale bar, 25 μm; close up, 10 μm.
(D) In transwell assays, SARS-CoV-2 is applied from the apical side to infect hiPS-BCECs (MOI 10). Mean ± SEM of n = 3 independent experiments, three technical replicates per condition. A significant increase in viral RNA was detected in the basolateral compartment by qRT-PCR. Two-way ANOVA with post hoc Sidak's multiple comparison test, ∗∗p = 0.001.
(E) Fluorescein transport study. The permeability coefficient is comparable 24 h post-infection for SARS-CoV-2 (MOI 10) or control-treated samples. Mean ± SEM from n = 3 independent experiments. Unpaired Student’s t test, p > 0.05.
(F) Host factors required for SARS-CoV-2 uptake are expressed in hiPS-BCECs and are diminished 24 h post-infection (MOI 10) compared with uninfected cells. Normalized to ACE2 in uninfected cells. Mean ± SEM from n = 3 individual experiments. Two-way ANOVA with post hoc Sidak's multiple comparison test, p > 0.05.
(G) Normalized expression of IFITM1 and IFITM2. Left: differential expression analysis of RNA-sequencing data with false discovery rate (FDR) correction for multiple comparisons. ∗p = 0.031 for IFITM1, p = 0.403 for IFITM2. Violin plots and mean of n = 6 independent experiments (uninfected) and n = 4 independent experiments (infected) are shown. Right: differential expression analysis of qRT-PCR data. Mean ± SEM of n = 2 experiments with independent virus isolates, three technical replicates per condition.
(H–K) TEM micrographs of SARS-CoV-2-infected hiPS-BCECs. (H) Overview of a TEM cross section of a hiPS-BCEC monolayer. After infection (MOI 10) from the apical side (top black arrow), virus is taken up, is evident in intracellular vesicles (middle black arrow), and is released from the cells on the basolateral side (bottom black arrow). (I and J) Detailed areas in higher resolution from (H). (I) Virus is evident in intracellular vesicles (black arrows). (J) Virus is released from the cells on the basolateral side (black arrow). (K) Neighboring hiPS-BCEC monocultures are connected by complex TJs constricting the paracellular space (black arrows). Furthermore, adhesion points (punctum adherens, black asterisk in K) anchored within the actin filament network were detected, indicating the integrity of cell-cell contacts. Scale bars as indicated.
(L) Representative immunofluorescence of SARS-CoV-2-infected hiPS-BCECs stained for SARS-CoV-2 N (red) and TJP1 (green) proteins show intact cell connectivity 24 h post-infection (MOI 10), counterstained by DAPI (blue). Scale bar, 50 μm.