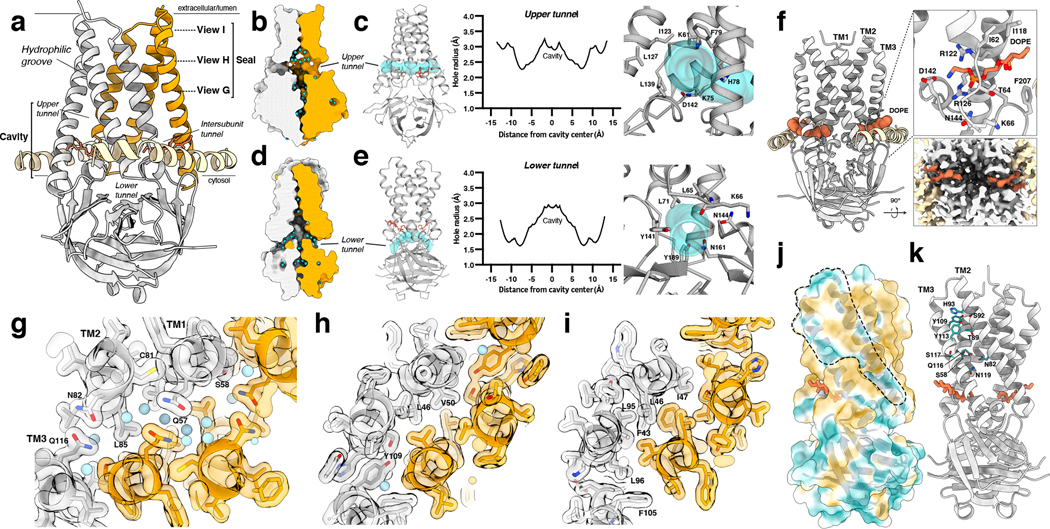
Figure 3. The 3a polar cavity and tunnels.
(a) View of a 3a dimer from the membrane plane with key regions labeled and with planes (dotted lines) and viewing direction (arrow) for (g-i) indicated. (b) The 3a upper tunnel with solvent-excluded surface shown for each subunit and water molecules shown as light blue spheres. The approximate lipid bilayer region is marked by black lines. One 3a subunit is colored orange and the other in gray. (c) (Left) Model of 3a (gray) with the HOLE path through the upper tunnel shown in transparent cyan. Lipid (coral) is displayed as sticks. (Middle) Radius of the upper tunnel as a function of distance from the cavity center calculated with HOLE. (Right) View into the upper tunnel from the membrane with key residues shown as sticks. (d) As in (b), for a view of the lower tunnel. (e) As in (c), for the lower tunnel, with the view on the right into the tunnel from the cytosol. (f) The intersubunit tunnel with bound DOPE lipid (coral) shown as space-filling spheres. (Right-top) Zoomed-in view of the lipid interaction with key residues shown as sticks. (Right-bottom) Cut-through view of the cryo-EM density of the lipid interaction region viewed from the cytosolic side. (g-i) Model and transparent surface of the viewing planes indicated in (a). Water molecules are shown as light blue spheres. One 3a subunit is colored orange and the other in gray. (j) The solvent excluded surface of 3a colored from hydrophilic (dark cyan) to most hydrophobic (dark orange). A membrane-facing hydrophilic groove is outlined. (k) Key residues in the hydrophilic groove are depicted as sticks colored in dark cyan.