Extended data figure 6. TET deficiency is associated with genome-wide accumulation of G-quadruplexes and R-loops.
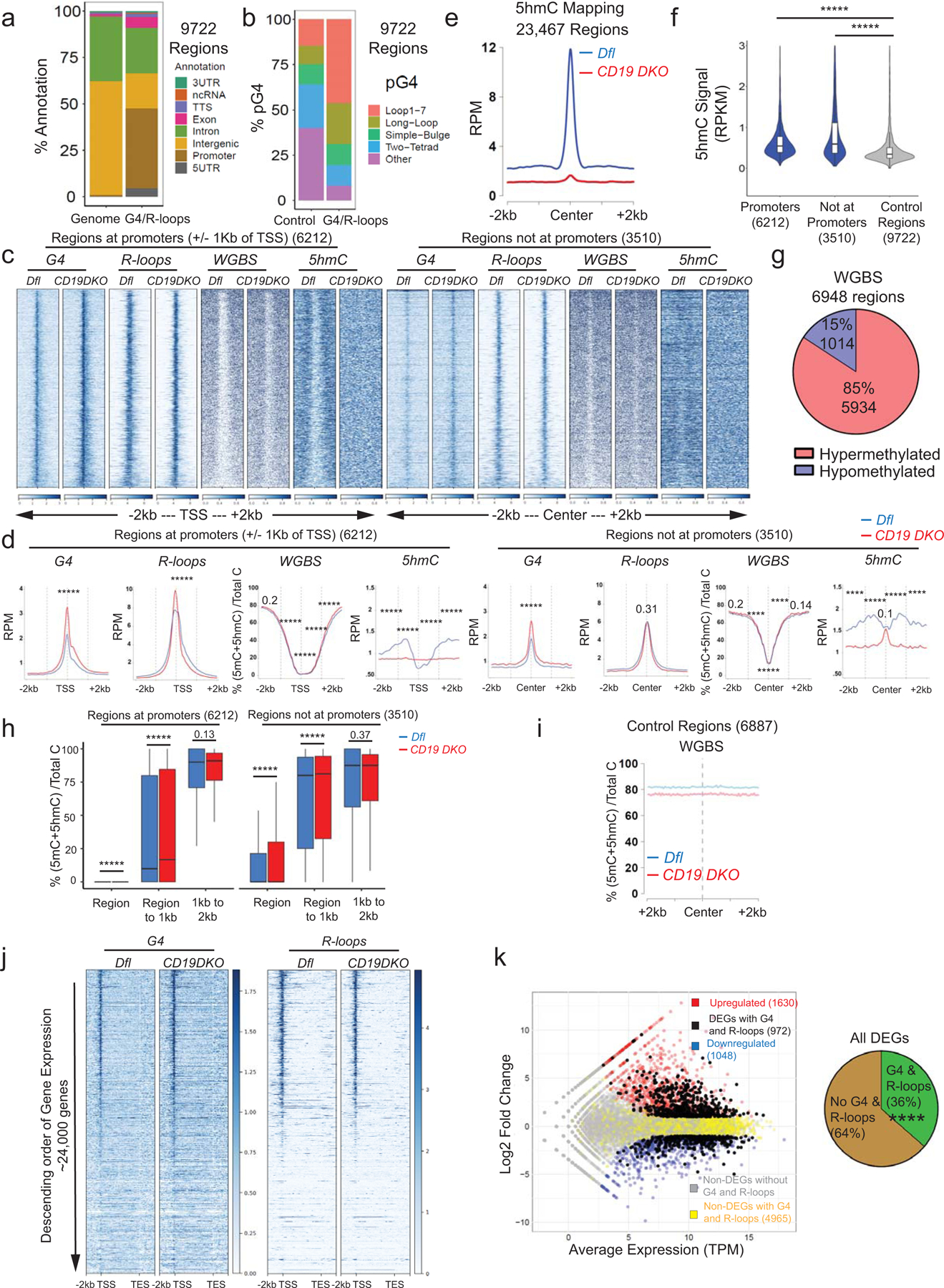
a) Genome annotations of regions enriched for G-quadruplexes (G4) and R-loops (right bar) compared to their representation in the mouse genome (mm10) (left bar). b) Relative representation of different classes of motifs predicted to form G-quadruplexes (pG4) in control regions selected randomly from the genome (left bar) and regions enriched for G-quadruplexes and R-loops (right bar). c) Heat maps showing enrichment (RPM) for G-quadruplexes and R-loops in CD19 DKO and control B cells. The signal is plotted in a +/− 2 kb window from the center of the regions ordered based on decreasing intensity from top to bottom in the entire 4 kb window. R-loop signal is plotted after background subtraction of MNase-alone control. d) Profile histograms showing the signals for G-quadruplexes (G4) (RPM, reads per million), R-loops (RPM), WGBS (percent of 5mC+5hmC/unmodified C) and 5hmC (RPM). The 9722 regions enriched for both G-quadruplexes and R-loops are divided into two categories – 6212 regions overlapping promoters (left panels) and 3510 regions not at promoters (right panels). Dashed grey lines indicate the center of the region and the 1 kb boundaries located on either side of the center. Blue and red lines show data from Dfl and CD19 DKO B cells, respectively. Asterisks represent statistical significance calculated by comparing the signals between Dfl and CD19 DKO B cells, either within the G-quadruplex and R-loop forming regions, the region to +/−1kb window or +/−1kb to 2kb window for respective datasets. e) Profile histograms showing the 5hmC signal in Dfl (blue) and CD19 DKO (red) B cells in 23,467 regions identified as enriched for 5hmC signal. f) Violin plots quantifying enrichment (RPKM) of 5hmC signal in Dfl B cells in the +/−1 kb from G-quadruplex and R-loop forming regions at promoters, non-promoter regions and control regions randomly located in euchromatin (Hi-C A genomic compartment) from 2 biological replicates. g) Pie chart showing the differentially methylated regions (DMRs) in CD19 DKO compared to control Dfl B cells. Of a total of 6948 DMRs identified by WGBS, 1014 (15%) showed reduced DNA methylation (hypomethylation) and 5934 (85%) showed increased DNA methylation (hypermethylation). h) Box and whisker plots quantifying percent of 5mC+5hmC/unmodified C (from WGBS) at and near the G4 and R-loop forming regions overlapping promoters and regions not overlapping promoters in Dfl (blue) and CD19 DKO (red) B cells from 2 biological replicates. The signal is plotted in three windows; window 1, within the G4 and R-loop regions; window 2, from region to +/− 1kb on either side and; window 3, +/−1kb to +/−2kb on either side. i) Percent of 5mC+5hmC/unmodified C (from WGBS) in random genomic regions of Dfl (blue) and CD19 DKO (red) B cells. j) Heatmaps of enrichment (RPM) for G-quadruplexes (left) and R-loops (right) in Dfl and CD19 DKO B cells, ordered in descending order of gene expression. k) MA plot (left) showing differentially expressed genes (DEGs) in CD19 DKO B cells. Red dots, upregulated DEGs; blue dots, downregulated DEGs; black dots, DEGs with G-quadruplexes and R-loops at their promoters (+/−1kb of TSS); yellow dots, non-DEGs with G-quadruplexes and R-loops at their promoters; grey dots, non-DEGs without G-quadruplexes and R-loops. The pie-chart (right) shows the percent of DEGs with (green) and without (brown) G-quadruplexes and R-loops at their promoters. Asterisks indicate statistical significance. Statistical significance is calculated using Kruskal-Wallis test and the ad hoc Dunn’s test in d), f) and h), Chi-square test in j). Boxes in box and whisker plots represent median (center) with 25th to 75 th percentile and whiskers represent maxima/minima. **** p value ≤0.0001 and ***** p value <0.000001.
