Figure 3.
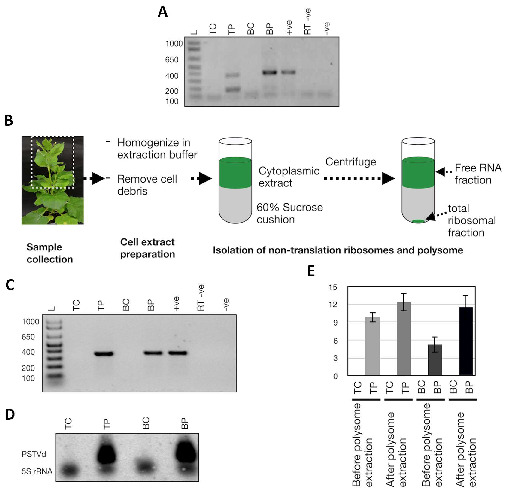
Detection of ribosome-associated PSTVd in host plants. Both Tomato cv. Rutgers and N. benthamiana plants were inoculated with PSTVdRG1. (A) Total RNA extracted and RT-PCR assay from these plants at 3 wpi was used to monitor the PSTVd infection. Lane L (Ladder); TC (tomato control), mock inoculated tomato plants; TP, PSTVdRG1 inoculated tomato plants; BC (N. benthamiana control), mock inoculated N. benthamiana plants; BP, PSTVdRG1-inoculated N. benthamiana plants; +ve, RT-PCR positive control; RT −ve, RT negative control and, −ve, PCR negative control. (B) Flow chart illustrating the details of the isolation of total ribosomes from leaf samples (see Materials and Methods). The resulting precipitates were subjected to RNA purification and analyzed by (C) RT-PCR and (D) Northern blot assays. The lanes were loaded as in (C). (E) RT-qPCR to evaluate the enrichment of PSTVdRG1 in the ribosomes. The expression change is presented on a log2 scale. Error bars indicate the standard deviation (SD).
