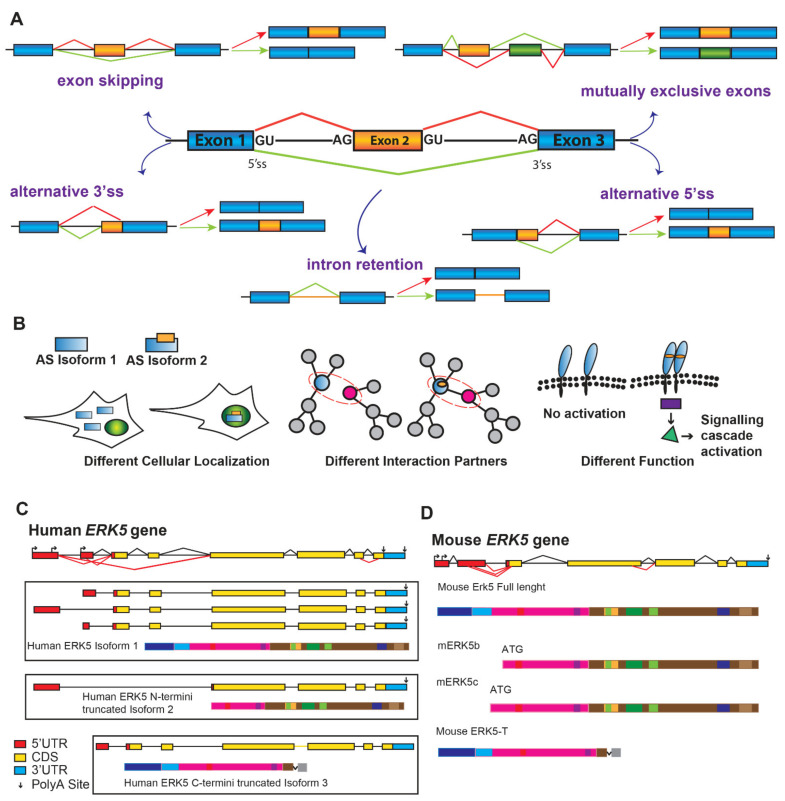
Figure 3.
ERK5 alternative splicing variants. (A). Schematic representation of five possible alternative splicing (AS) mechanisms. Constitutive exons = blue, alternative exons = orange, introns = lines. Different AS mRNAs result from exon skipping, intron retention, usage of alternative 3′or 5′splice sites (3′ ss and 5′ ss), and from the selection of mutually exclusive exons. (B). Protein isoforms generated through AS reaction can differ for their cellular localization (cytoplasmic vs. nuclear in the case of isoform 1 and 2) (left), for their protein-protein interactions (center), or their function as components of a signaling pathway (right). (C). Predicted transcripts for human ERK5 gene based on supported EST adapted from ASPicDB (http://srv00.recas.ba.infn.it/ASPicDB/, accessed on 7 January 2022). The exon-intron scheme is depicted with 5′UTR in red, the coding region (CDS) in yellow, and 3′UTR in light blue; polyadenylated transcripts are marked with vertical arrows; alternative transcription start sites are also represented. The canonical ERK5 human protein, generated by three different transcripts that differ in their 5′UTR (exons in red), is shown as isoform 1. Another mRNA transcript generated by skipping of first exons can encode for a protein isoform deleted of the N-termini region (isoform 2), whereas the ERK5-C termini truncated protein, generated by intron retention, lacks the cytoplasmic targeting and MEK5 binding domains (isoform 3). (D). Schematic representation of murine ERK5 gene with 5′UTR in red, CDS in yellow, and 3′UTR in light blue. Three different ERK5 murine proteins are depicted: mERK5b and c differ for their ATG and lack the cytoplasmic targeting domain (blue), mouse ERK5-T lacks the NLS and the PR2 domain.