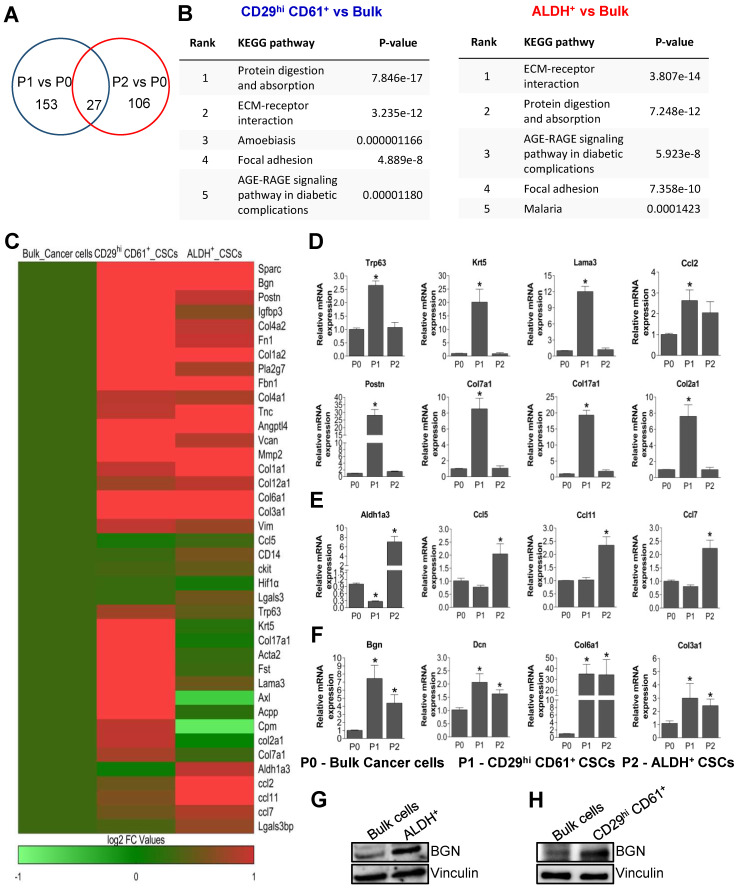
Figure 1.
Upregulation of BGN in BCSCs derived from PyMT tumor cells. (A) Venn diagram depicting the upregulated genes in CD29hi CD61+ (P1) and ALDH+ (P2) BCSCs as well as overlapping genes in both BCSCs as compared to bulk cells (P0). (B) KEGG pathway analysis showed that ECM receptor interaction and focal adhesion pathways were commonly upregulated in both BCSCs. (C) Heat map data showing the top 40 candidates identified from RNA sequencing. qRT-PCR data showing validation of genes in BCSCs, CD29hi CD61+ (P1) and ALDH+ (P2) as compared to bulk cells, (D) expression of ECM genes, (E) BCSCs marker expression, (F) ECM genes upregulated in BCSCs. Immunoblots depicting expression of BGN in sorted BCSCs from MMTV-PyMT tumor cells (FF99), (G) ALDH+ and (H) CD29hi CD61+ compared to bulk cells. Data shown as mean ± SEM of three independent experiments. Statistical significance was determined using two-tailed t-test; * p ≤ 0.05 as compared to bulk cells.