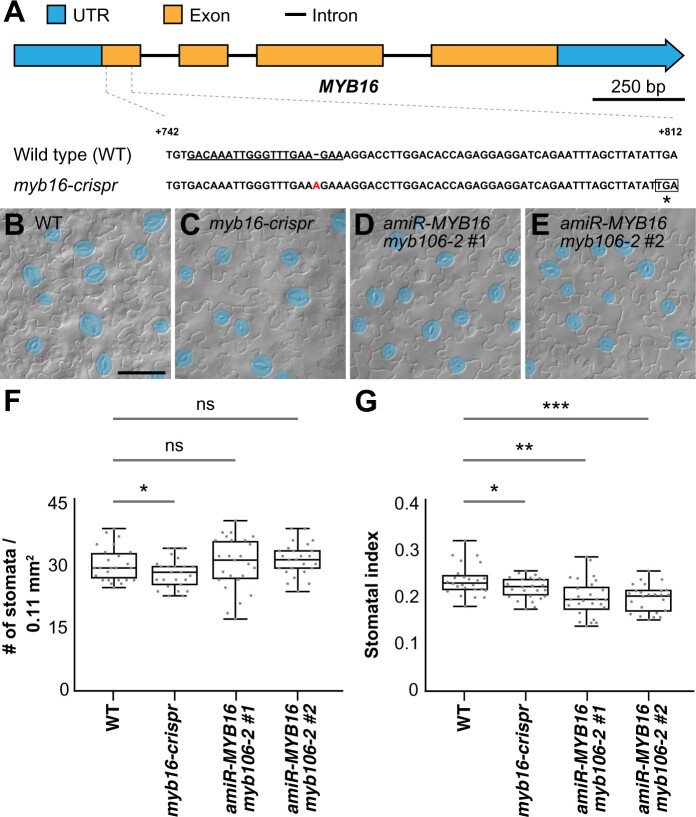
Figure 2.
The stomatal index is reduced in MIXTA-like loss-of-function mutants. A, Scheme of the MYB16 gene with the targeted sequence of CRISPR guide RNA (underlined). An adenine (A) insertion results in a premature stop codon (asterisk). B–E, DIC images of lower epidermis from 10-dpg WT true leaves (B), myb16-crispr (C) and two independent lines in which MYB16 artificial micro-RNA (amiR-MYB16) was introduced into the myb106-2 background (D) and (E). Mature GCs are pseudo-colored in blue. Scale bars, 50 µm. F, Quantification of stomatal density in (B) to (E). Stomatal density was reduced in the clustered regularly interspaced short palindromic repeats (CRISPR)-edited MYB16 line (myb16-crispr) but not in amiR-MYB16/myb106-2. n = 25 seedlings. *P < 0.05, by Wilcoxon signed-rank test. Data are medians (interquartile range); ns, not significant. G, Quantification of stomatal index in (B–E). Stomatal index was reduced in both myb16-crispr and amiR-MYB16/myb106-2. n = 25 seedlings. *P < 0.05; **P < 0.01; ***P < 0.001, by Wilcoxon signed-rank test. Data are medians (interquartile range).