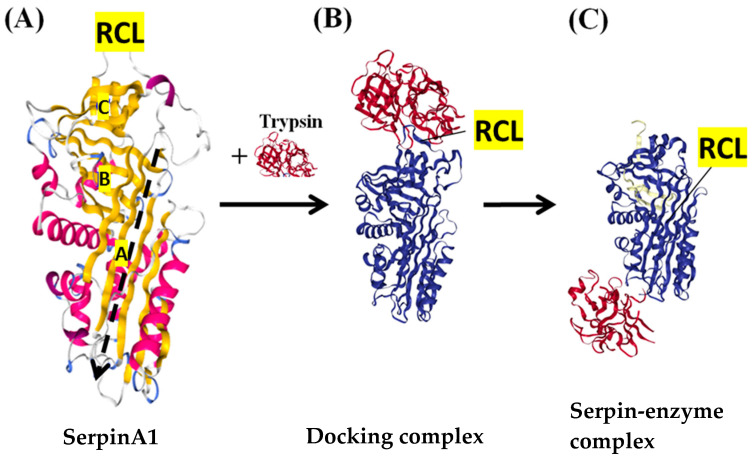
Figure 1.
The structure and mechanism of inhibitory serpins. (A) The native structure of the archetypal serpin—serpinA1 (protein data bank (PDB) code 1QLP). The α-helices are shown in pink, and 3 β-sheets with gold color are marked with A, B and C. The reactive center loop (RCL) is at the top of the molecule and between A sheet and C sheet. The black dashed line indicates the path of RCL insertion after RCL is cleaved by the target protease. (B) The docking complex between serpinA1 (blue) and inactive trypsin (red) (PDB code 1OPH). The protease docked onto the RCL. (C) The final serpin–enzyme complex (PDB code 1EZX). After the RCL is cut, the serpin undergoes a transition from the stressed form to the relaxed form, and the docking complex becomes the serpin–enzyme complex, in which the distorted protease hangs at the base of the serpin molecule.