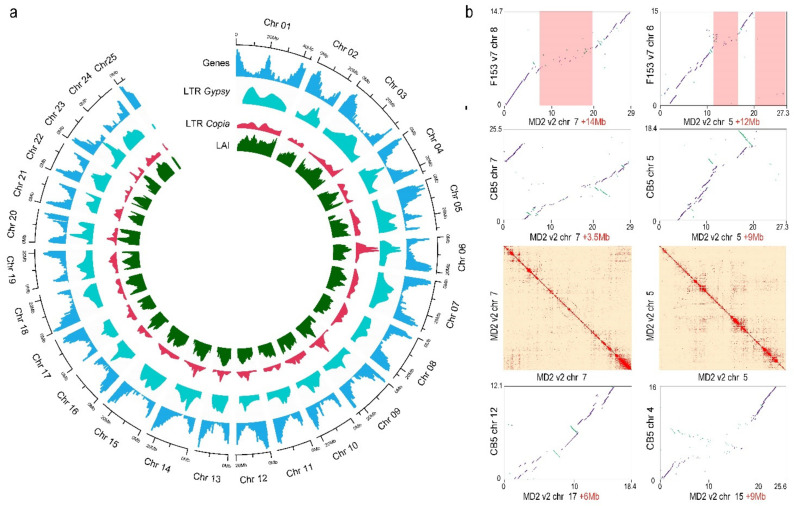
Figure 1.
MD2 v2 P0 genomic features’ landscape and comparative analysis. (a) CIRCOS plot illustrating the location/density of gene models, Ty3/Gypsy LTR, Ty1/Copia LTR, and LTR Assembly Index (LAI) score along the MD2 v2 P0 assembled chromosomes; (b) Visual representation of the quality of the MD2 v2 genome compared to other high-quality pineapple genomes (F153 v7 and CB5). Numbers in the x and y axes represent Mb sequences. Additional sequences assembled in MD2 v2 were primarily located in regions with high amounts of repeats (i.e., centromeres, highlighted with red shaded boxes). Contiguous interaction along the Hi-C plot validates the quality of the newly assembled regions. In each chromosome comparison, the number highlighted in red indicates the additional sequences assembled in the MD2 v2 assembly.