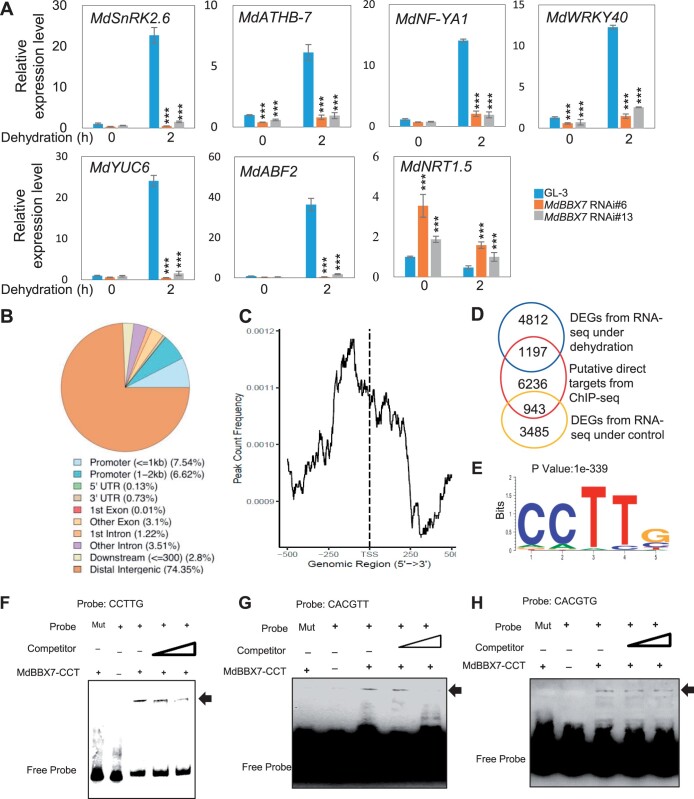
Figure 3.
Identification of MdBBX7 recognition sites. A, Expression of drought-responsive genes in MdBBX7 RNAi plants under control and dehydration conditions. B, Distribution of MdBBX7 binding regions in the apple genome. ChIP was performed using tissue-cultured GL-3 seedlings with anti-MdBBX7 antibody. C, Peak distance from the TSS of MdBBX7. The peaks were highly enriched from −250 bp to 0 from the TSS. D, Numbers of regulon detected by ChIP-seq and RNA-seq under control and dehydration conditions. DEGs were from the MdBBX7 RNAi plants detected by RNA-seq under control and dehydration conditions. E, The CCTTG element was identified from ChIP-seq analysis using MEME-ChIP. F–H, EMSA analysis showing that MdBBX7 protein bound to the element of CCTTG (F), CACGTT (G), and CACGTG (H). The C terminal region of MdBBX7 containing the CCT domain (207–410 aa) was used. Mut was presented the mutant probe. Binding sites in mutant probe were replaced with AAAAA or AAAAAA. Data are means ± sd (n = 3). Asterisks indicated values significantly different between the transgenic lines and the GL-3 in each group (0 h and 2 h). One-way ANOVA (Tukey’s test) was performed and statistically significant differences were indicated by ***P < 0.001.