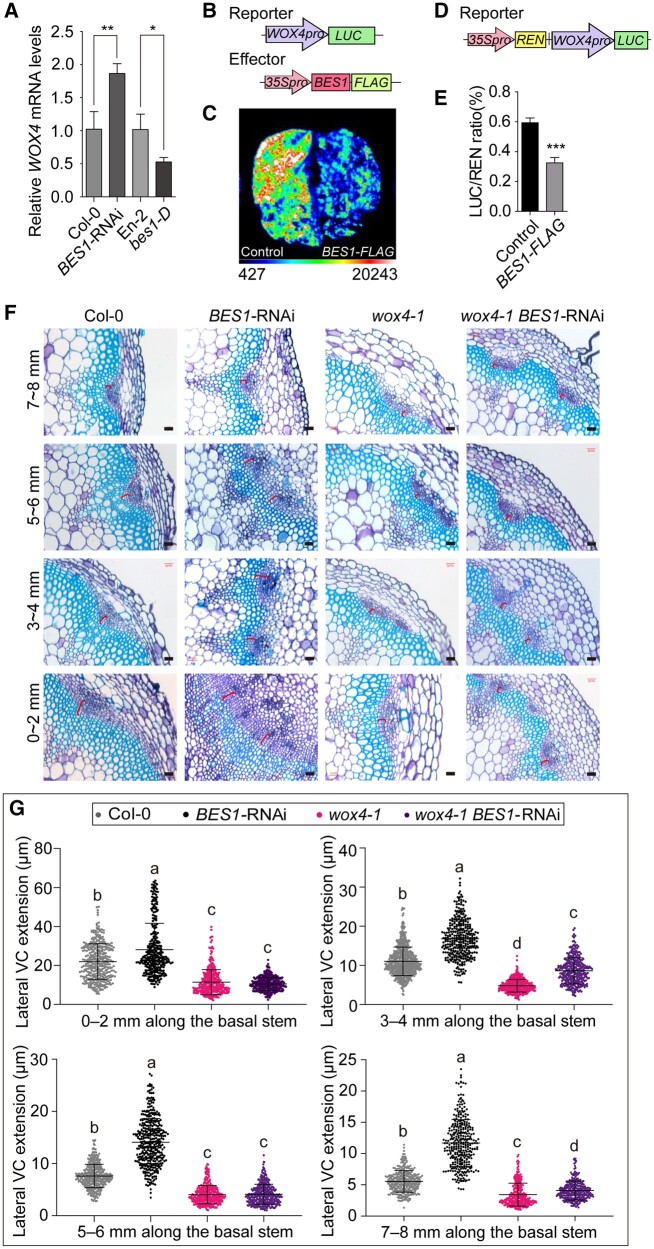
Figure 3.
BES1 inhibits cambial cell proliferation by repressing WOX4 transcription. A, Relative expression levels of WOX4 in Col-0, BES1-RNAi, En-2, and bes1-D. Data are means ± sd (n = 3). P values were determined by Student’s t test; **P < 0.01; *P < 0.05. B, Diagrams representing the luciferase reporter and effector constructs used in the luciferase assays (C). The WOX4 promoter drives the expression of the firefly LUC reporter. The BES1-FLAG effector construct is driven by the 35S promoter. We used 35Spro:GFP as a negative control. C, Transient luciferase assays in N. benthamiana leaves demonstrate the repression of WOX4pro:LUC activity by co-expression with BES1-FLAG. D, Diagram of the reporter used for the dual bioluminescence assay (E). The reporter consists of the WOX4pro:LUC cassette placed downstream of 35Spro:REN. E, Dual-luciferase assays in N. benthamiana leaves confirm the repression of luciferase activity from the WOX4pro:LUC reporter by the BES1-FLAG effector. Data are means ± sd (n = 3). P values were determined by Student’s t test. ***P < 0.001. F, Representative cross sections illustrating the extension of lateral VC in the basal stems of Col-0, BES1-RNAi, wox4-1, and wox4-1 BES1-RNAi plants. A segment of 8 mm was collected at the base of the stem immediately above the shoot–hypocotyl junction and cut into four equal parts, marked as 0–2, 3–4, 5–6, and 7–8 mm. The red bracket indicates the measured areas. Bars represent 20 μm. G, Quantification of lateral VC extension in (F) by ImageJ. Data are means ± se. n > 300 for each sample were analyzed using ANOVA with post hoc Tukey’s test. Means with a P value ≤ 0.05 were considered statistically different among multiple comparisons. Distinct letters (a–c) indicate statistically significant difference among all samples.