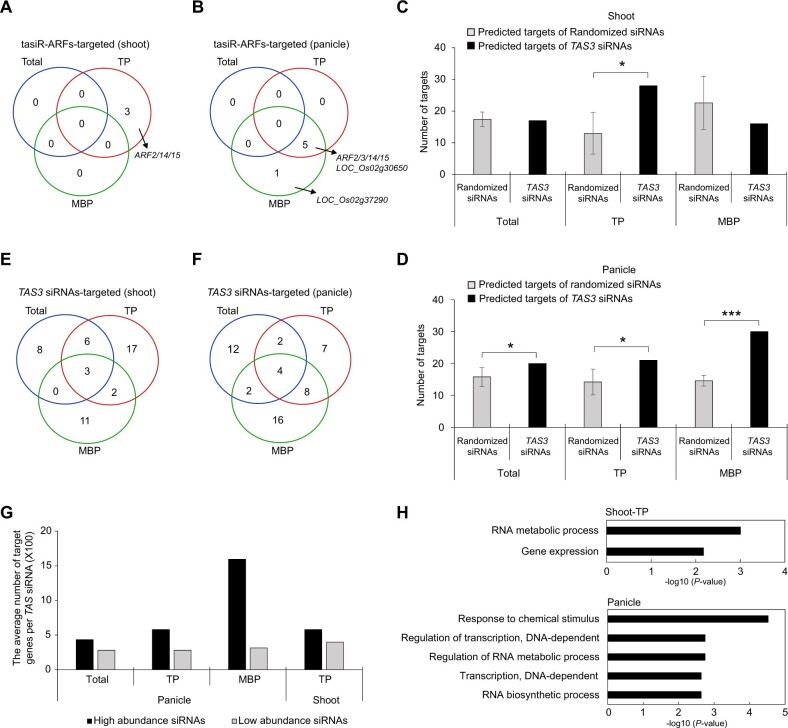
Figure 4.
Identification of TAS3 siRNA target genes in rice. A and B, Numbers of tasiR-ARF target genes as determined by PARE analysis for input (Total), TP, and MBP samples from rice shoots (A) and panicles (B). C and D, Numbers of predicted targets of randomized siRNAs and TAS3 siRNAs in Total, TP, and MBP samples from shoots (C) and panicles (D). E and F, Numbers of TAS3 siRNA target genes as determined by PARE analysis for Total, TP, and MBP samples from rice shoots (E) and panicles (F). G, Summary for TAS3 siRNA targets in panicle Total, TP, and MBP and shoot TP samples. The black and gray rectangles represent target transcripts cleaved by TAS3 siRNAs with RPM ≥1 and TAS3 siRNAs with RPM <1, respectively. H, Enriched GO terms for TAS3 siRNA target genes in panicle Total, TP, and MBP and shoot TP samples as determined with the online tool agriGO v2.0 (http://systemsbiology.cau.edu.cn/agriGOv2/). The cutoff parameters for the identification of target genes were “degradomic category 0” and “P-value ≤0.05.” In (C and D), significant differences as compared to the randomized group are denoted by *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, respectively, with the P-values being derived from one-sample test (95% confidence interval). Number of target genes is displayed as mean ± standard deviation of five biological repeats.