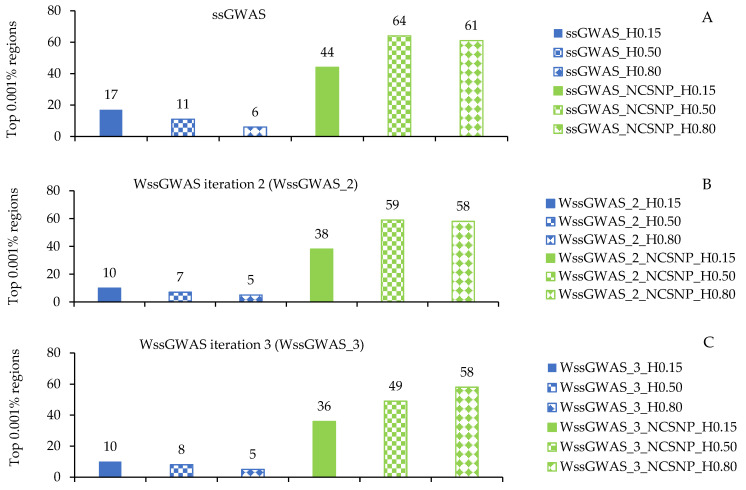
Figure 1.
Number of top 0.001% genomic regions for yearling temperament in American Angus cattle found by non-weighted single-step GWAS (ssGWAS) (A) and weighted ssGWAS (WssGWAS) in the second (B) and third (C) iterations. H0.15, H0.50, and H0.80: only haplotypes from blocks with linkage disequilibrium (LD) thresholds of 0.15, 0.50, and 0.80, respectively; NCSNP_H0.15, NCSNP_H0.50, and NCSNP_H0.80: non-clustered SNPs (NCSNP) and haplotypes from blocks with LD thresholds of 0.15, 0.50, and 0.80, respectively. The column colors highlight not including (blue) or including NCSNP (green). The column filling highlights different LD thresholds (0.15, 0.50, and 0.80 with a solid, square, and diamond filling, respectively).