FIGURE 6.
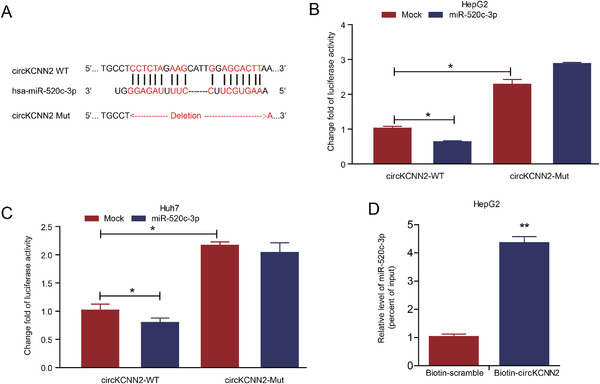
Confirmation of the binding between miR‐520c‐3p and circKCNN2. (A) The predicted binding sites between miR‐520c‐3p and circKCNN2. (B, C) The luciferase activities in HepG2 and Huh7 cells after co‐transfection with the reporter plasmids and miR‐520c‐3p mimics. Mock, miRNA negative control; miR‐520c‐3p, mimics of miR‐520‐3p. The group of circKCNN2‐WT + Mock served as a reference to calculate change fold value. (D) The amount of miR‐520c‐3p in the RNA pulled down by using the biotin‐circKCNN2 probe. The group of biotin‐scramble served as a reference for calculating relative value. All the assays were performed at least three times. N = 3 for each group. *P < 0.05, **P < 0.01, ***P < 0.001
