Figure 1. Flow chart for exome-by-phenome-wide association analysis using electronic health record phenotypes.
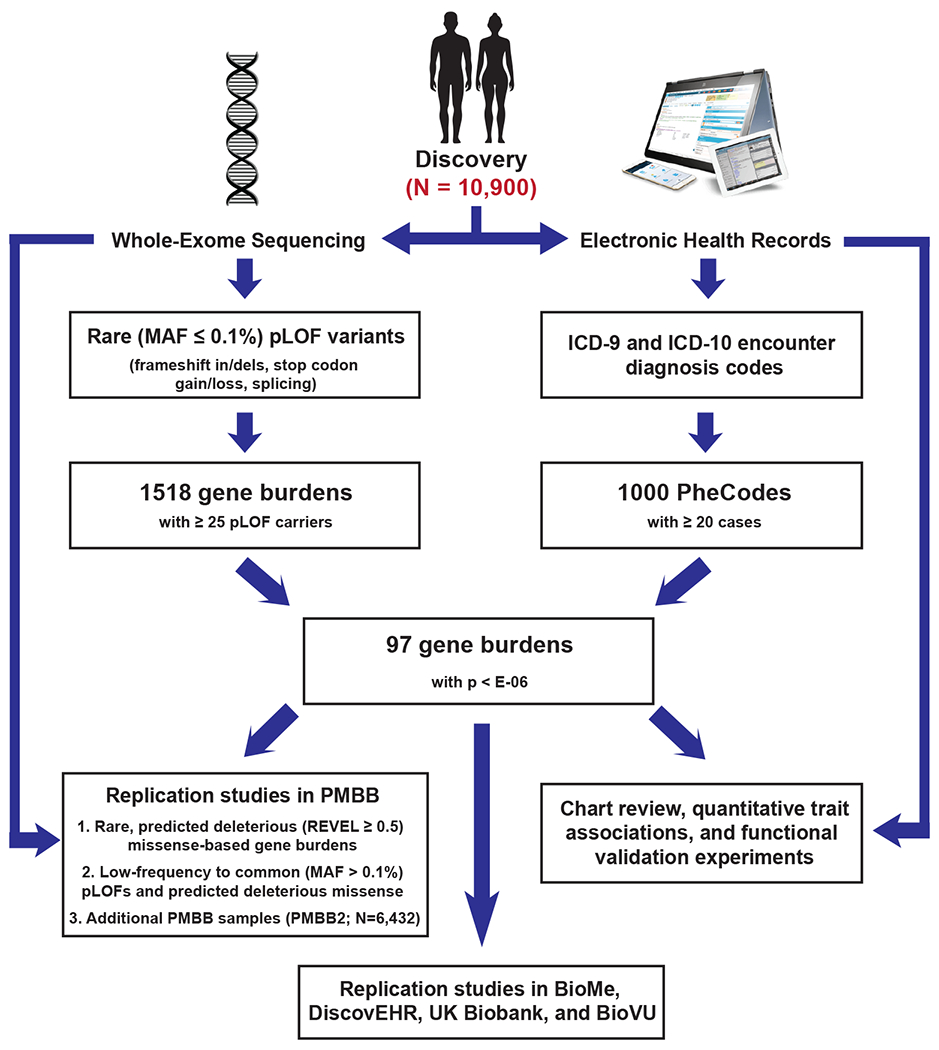
Flowchart diagram outlining the primary methodologies used for conducting the exome-by-phenome-wide association study and for evaluation of the robustness of the associations, indicating that 97 genes had associations at a significance level of p<E-06 via logistic regression. The pathways starting with short descending arrows represent the 'discovery phase', in which predicted loss-of-function (pLOF)-based gene burdens were studied on an exome-by-phenome-wide scale in 10,900 individuals from the Penn Medicine Biobank (PMBB). “Replication studies in PMBB” refers to analyses of gene-phenotype associations using REVEL-informed missense-based gene burdens and univariate analyses within the discovery PMBB cohort, as well as in an independent cohort of African Americans in the PMBB (the PMBB2 cohort; N=6,432). Additional replication studies included analyses of gene-phenotype associations using pLOF-based gene burdens, REVEL-informed missense-based gene burdens, and univariate analyses in BioMe (N=23,989), DiscovEHR (N=85,450), and the UK Biobank (N=32,268), as well as univariate analyses in BioVU (N=66,400).
