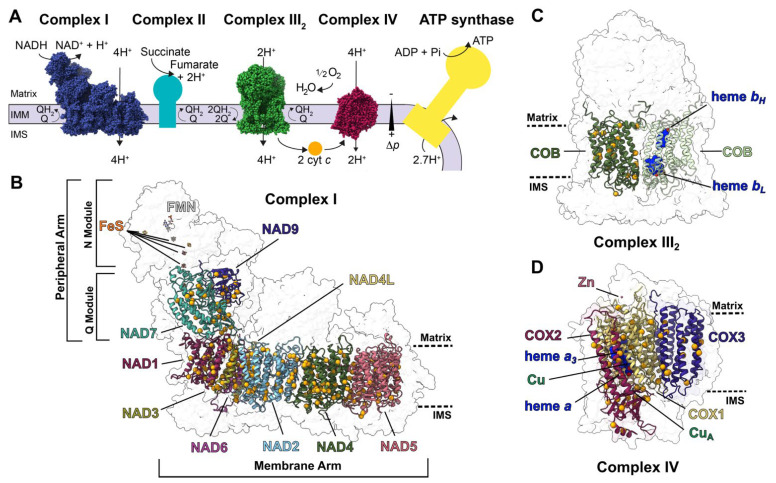
Figure 1.
RNA-edited subunits of plant respiratory complexes: (A) Schematic overview of the plant canonical respiratory chain. Complexes I-IV and ATP synthase are shown in the inner mitochondrial membrane (IMM). Complex I (blue, PDB: 7AR8), complex III2 (green, PDB: 7JRG) and complex IV (magenta, PDB: 7JRO) are shown with their atomic structures in sphere representation [11,12]. Complexes without structures (complex II, teal; ATP synthase, yellow) are represented in boxes. Substrates and products (NADH, NAD+, succinate, fumarate, O2, H2O, ADP, Pi, ATP), electron carriers (quinone, Q; quinol, QH2; cytochrome c, cyt c), proton pumping stoichiometry (H+), protonmotive force (∆p), matrix, intermembrane space (IMS) and inner mitochondrial membrane (IMM) are indicated. (B–D) Subunits of complex I (B), complex III2 (C) and complex IV (D) that undergo RNA editing are shown in colored cartoons. Edited residues are shown as orange spheres overlayed over the transparent surface of the complex. Approximate locations of the matrix and IMS are shown in dashed lines. (B) In addition to the edited subunits, complex I’s flavin mononucleotide (FMN) and iron-sulfur (FeS) co-factors are shown in stick representation. Complex I’s membrane and peripheral arms and approximate locations of the N (NADH-binding) and Q (quinone-binding) modules are marked. (C) Both COB subunits of complex III2 are shown. Dark-green COB highlights the edited residues. Light-green COB highlights the subunit’s heme bH and bL (dark blue spheres). (D) Complex IV’s heme a, heme a3, Zn2+, copper-A (CuA) co-factors are shown in spheres.