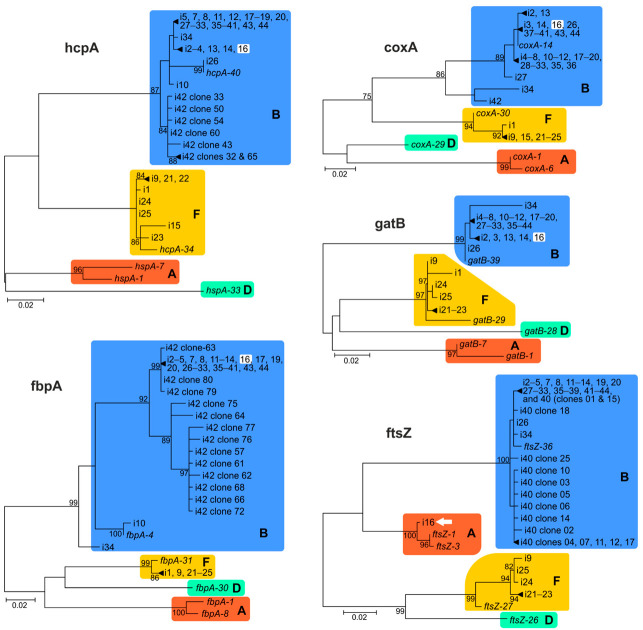
Figure 1.
Maximum likelihood (ML) phylogenetic trees for each MLST gene. Acrididae isolates (Table 2), supergroups (A, B, F, and D), and bootstrap values are indicated. Model of nucleotide substitutions T92+G was used for gatB, hcpA, and fbpA datasets; HKY+G for coxA; T92+G for ftsZ. White squares and the arrow indicate a case (i-16) of a supergroup clustering conflict. See original files in Supplementary Materials.