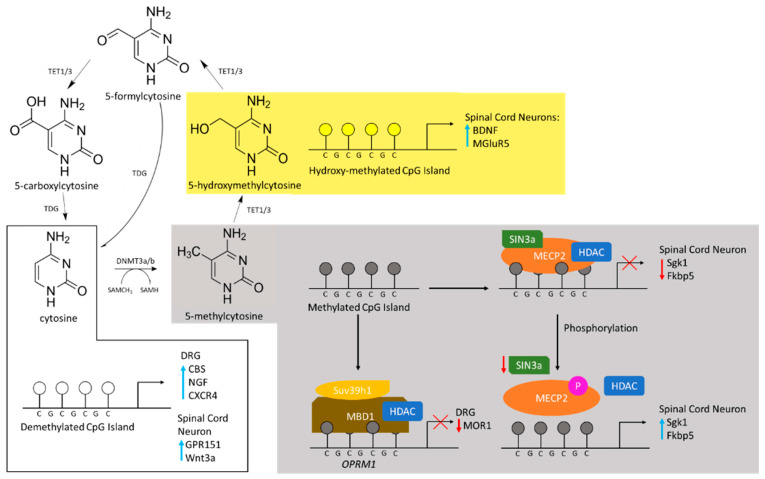
Figure 2.
Summary of the cytosine methylation cycle, readers of DNA methylation, and mechanisms of gene expression and inhibition. Demethylated cytosine is methylated by DNMT3a and 3b using the methyl group donated by the cofactor SAM. 5-methylcytosine can be hydroxylated to 5-hydroxymethylcytosine by TET enzymes and then further oxidised to 5-carboxylcytosine. Thymine DNA glycosylase (TDG) may convert 5-carboxylcytosine or 5-formylcytosine back to cytosine. Demethylated CpG islands are associated with the upregulation of various proteins in various pain models (white box). Within the dorsal root ganglia, demethylation is associated with upregulation of CBS, NGF and CXCR4. In spinal cord neurons, demethylation is associated with the transcription of GPR151 and Wnt3a. Different preclinical models show pain is associated with methylated CpG islands (grey box). In dorsal root ganglia, methylation may be read by MBD1, which indirectly leads to downregulation of the μ-opioid receptor 1 (MOR1). Under normal conditions within spinal cord neurons, MeCP2 may bind and recruit SIN3a and the HDAC to repress transcription of Sgk1 and Fkbp5. In a CFA model, the binding of MeCP2 is reversed by phosphorylation and leads to the transcription of Sgk1 and Fkpb5, which are associated with mechanical hypersensitivity. TET-dependent demethylation of 5-methylcytosine is associated with the transcription of BDNF and MGluR5 via hydroxyl-methylated promoter regions of spinal cord neurons (yellow box).