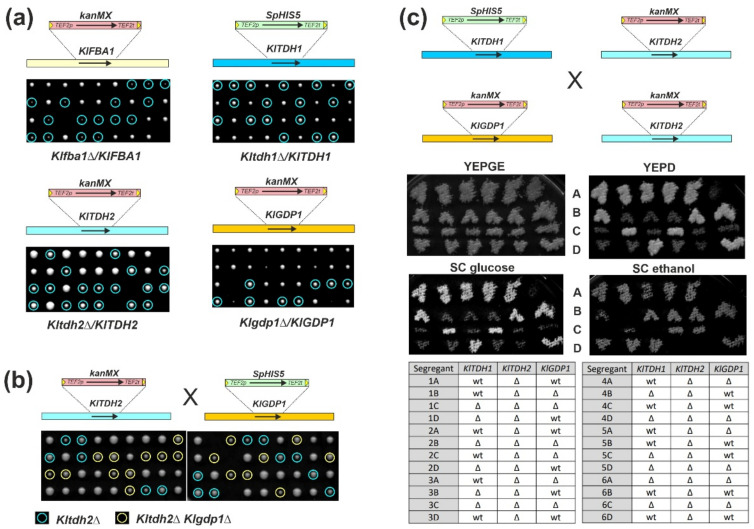
Figure 1.
Tetrad analyses of deletion mutants. (a) Strains with heterozygous deletions of the genes indicated in the schematic drawings were sporulated and subjected to tetrad analyses on rich medium (YEPD). After micromanipulation, plates were incubated for 2 days (images with smaller colonies) or 3 days (Kltdh2/KlTDH2) at 30 °C. Growth was documented by scanning the plates and adjustment of brightness using Corel Photo-Paint. Nine representative tetrads from a total of at least eighteen separated from each diploid are shown. Light-blue circles mark the segregants carrying the respective deletions, as judged from replica plating onto selective marker plates. Note that in the case of the heterozygous diploid for the Klgdp1 deletion, another deletion, Kltrp1::kanMX, carried the same marker cassette. Therefore, only segregants unequivocally carrying the deletion, i.e., those that were both prototrophic for tryptophan and resistant to G418, are highlighted. Strains employed were KHO483 and KHO505 (top row), and KHO455 and KHO461 (bottom row). (b) Tetrad analysis of a heterozygous diploid (KHO476) obtained from crossing a Kltdh2::kanMX deletion with a Klgdp1::SpHIS5 deletion. Segregants carrying a single Kltdh2 deletion are highlighted with a light-blue circle. Double deletions are indicated by yellow circles. The plate was incubated for 3 days at 30 °C after micromanipulation and all 18 tetrads separated are shown. (c) Tetrad analysis of a diploid strain heterozygous for both the Kltdh1 and the Klgdp1 deletion, but homozygous for the Kltdh2 deletion (KHO506). After micromanipulation, tetrads were allowed to grow on rich medium with 2% glycerol and 2% ethanol as carbon sources (YEPGE) for 3 days at 30 °C. Segregants were picked onto a master plate of the same medium and allowed to grow overnight. Numbers refer to the tetrad analyzed and signs from top to bottom designate segregants A to D as indicated. Only 6 representative tetrads from a total of 14 tetrads with 4 viable segregants are shown. Growth was documented after replica plating onto the indicated media and incubation over night at 30 °C. The relevant genotype for each segregant can be deduced from the table given below. Segregants carrying Kltdh1 deletions in combination with Kltdh2 and/or Klgdp1 were assigned from their ability to grow on synthetic medium lacking histidine and with 2% glycerol (w/v) as a sole carbon source. Klgdp1 deletions were assigned by PCR using flanking oligonucleotides as primers. SC = synthetic complete medium with either 2% glucose (w/v) or 2% ethanol (v/v).