Abstract
Background
A range of safe and effective vaccines against SARS CoV 2 are needed to address the COVID 19 pandemic. We aimed to assess the safety and efficacy of the COVID-19 vaccine SCB-2019.
Methods
This ongoing phase 2 and 3 double-blind, placebo-controlled trial was done in adults aged 18 years and older who were in good health or with a stable chronic health condition, at 31 sites in five countries (Belgium, Brazil, Colombia, Philippines, and South Africa). The participants were randomly assigned 1:1 using a centralised internet randomisation system to receive two 0·5 mL intramuscular doses of SCB-2019 (30 μg, adjuvanted with 1·50 mg CpG-1018 and 0·75 mg alum) or placebo (0·9% sodium chloride for injection supplied in 10 mL ampoules) 21 days apart. All study staff and participants were masked, but vaccine administrators were not. Primary endpoints were vaccine efficacy, measured by RT-PCR-confirmed COVID-19 of any severity with onset from 14 days after the second dose in baseline SARS-CoV-2 seronegative participants (the per-protocol population), and the safety and solicited local and systemic adverse events in the phase 2 subset. This study is registered on EudraCT (2020–004272–17) and ClinicalTrials.gov (NCT04672395).
Findings
30 174 participants were enrolled from March 24, 2021, until the cutoff date of Aug 10, 2021, of whom 30 128 received their first assigned vaccine (n=15 064) or a placebo injection (n=15 064). The per-protocol population consisted of 12 355 baseline SARS-CoV-2-naive participants (6251 vaccinees and 6104 placebo recipients). Most exclusions (13 389 [44·4%]) were because of seropositivity at baseline. There were 207 confirmed per-protocol cases of COVID-19 at 14 days after the second dose, 52 vaccinees versus 155 placebo recipients, and an overall vaccine efficacy against any severity COVID-19 of 67·2% (95·72% CI 54·3–76·8), 83·7% (97·86% CI 55·9–95·4) against moderate-to-severe COVID-19, and 100% (97·86% CI 25·3–100·0) against severe COVID-19. All COVID-19 cases were due to virus variants; vaccine efficacy against any severity COVID-19 due to the three predominant variants was 78·7% (95% CI 57·3–90·4) for delta, 91·8% (44·9–99·8) for gamma, and 58·6% (13·3–81·5) for mu. No safety issues emerged in the follow-up period for the efficacy analysis (median of 82 days [IQR 63–103]). The vaccine elicited higher rates of mainly mild-to-moderate injection site pain than the placebo after the first (35·7% [287 of 803] vs 10·3% [81 of 786]) and second (26·9% [189 of 702] vs 7·4% [52 of 699]) doses, but the rates of other solicited local and systemic adverse events were similar between the groups.
Interpretation
Two doses of SCB-2019 vaccine plus CpG and alum provides notable protection against the entire severity spectrum of COVID-19 caused by circulating SAR-CoV-2 viruses, including the predominating delta variant.
Funding
Clover Biopharmaceuticals and the Coalition for Epidemic Preparedness Innovations.
Introduction
After the emergence of SARS-CoV-2 in Wuhan, China,1 and the subsequent global dissemination, there have been almost 265 million COVID-19 cases resulting in 5·2 million deaths by Dec 3, 2021.2 Major research efforts have resulted in more than 130 vaccines in clinical development, most targeting the spike glycoprotein (S protein), the main viral protein that induces neutralising antibodies,3 with almost 10 billion doses of these vaccines administered to date.2, 4 Widely used vaccines with shown efficacy include inactivated whole-virus vaccines,5, 6 mRNA vaccines coding for the S protein encapsulated in lipid nanoparticles,7, 8 or viral vectors expressing the S protein.9, 10 An adjuvanted, recombinant S protein has also been shown to have efficacy but is not yet authorised for use.11 Clover Biopharmaceuticals has developed a vaccine candidate, SCB-2019, consisting of a recombinant SARS-CoV-2 S protein stabilised in the native prefusion trimeric conformation using its proprietary Trimer-Tag technology.12 Preclinical studies have shown that adjuvanted SCB-2019 elicits protective neutralising antibody responses against SARS-CoV-2 challenge in non-human primates.13 A phase 1 study in adults showed robust immune responses with SCB-2019 when adjuvanted with the toll-like receptor agonist CpG-1018 combined with alum,14 with antibodies persisting at more than baseline for 6 months after vaccination.15 We report here the results from the phase 2 and ongoing phase 3 Study evaluating Protective Efficacy and safety of Clover's Trimeric Recombinant protein-based and Adjuvanted COVID-19 vaccine (SPECTRA). These studies are being done in adults with no evidence of previous exposure to SARS-CoV-2 to obtain data on the safety and tolerability of two doses of SCB-2019 plus CpG-1018 and alum and their efficacy in preventing RT-PCR-confirmed symptomatic COVID-19.
Research in context.
Evidence before this study
The massive global effort to develop effective SARS-CoV-2 vaccines to combat COVID-19 has led to an unprecedented literature database of preclinical and clinical studies of vaccine candidates. An unrestricted PubMed search on Nov 25, 2021, with the terms “COVID-19”, “SARS-CoV-2”, “vaccine”, and “efficacy” produced 2177 results, which was refined to 30 by the addition of “phase 3 clinical trial”. These references generally describe the results of several clinical trials of currently authorised COVID-19 vaccines, which are now in use globally. The main focus of these vaccines is the SARS-CoV-2 spike (S)-protein. In most cases, the S protein antigen is targeted using mRNA and human or chimpanzee adenovirus-vectored mRNA coding for it, but one vaccine uses an adjuvanted form of the full-length S glycoprotein of the prototype (Wuhan) strain. One publication has identified a statistically significant correlation between the levels of S protein binding antibodies in vaccinees, measured in one laboratory and standardised with an international standard serum pool, and the proven clinical efficacies of four authorised vaccines. The global requirement for effective COVID-19 vaccines has not yet been met, and the storage requirements during distribution and use of some of the available vaccines means new vaccines are still needed.
Added value of this study
We have previously reported that an S protein subunit vaccine against SARS-CoV-2 (SCB-2019) consisting of the trimeric structure of the S protein adjuvanted with CpG-1018 and alum elicits a robust immune response in adults 14 days after a second dose. This study confirms that this immune response is effective in protecting against any severity of COVID-19 in adults, and is highly effective against moderate-to-severe and severe disease and admission to hospital due to COVID-19. Furthermore, this study was done when the landscape of circulating SARS-CoV-2 had changed substantially since the first vaccines were assessed, with the prototype virus being essentially replaced by variants, notably delta, gamma, and mu. It is reassuring to note that SCB-2019 has demonstrated efficacy against each of these variants. Further, the reactogenicity profile of SCB-2019, with 36·4% of the first dose and 28·1% of the second dose associated with solicited adverse events, was markedly better than that reported for the authorised vaccines.
Implications of all the available evidence
In high-income countries there are high rates of vaccination coverage with effective COVID-19 vaccines, but vaccination programmes in lower-income nations are lagging in part because of the high cost and low availability of suitable vaccines. Notably, the storage requirements of mRNA vaccines, which require very low temperatures, impose notable logistical difficulties in the distribution and use of such vaccines in many low-income countries. The evidence from this study shows that SCB-2019 is effective, particularly against moderate-to-severe COVID-19, with a superior reactogenicity profile and is kept at normal refrigerator temperatures. Once authorised this vaccine will be a welcome addition to the armamentarium against COVID-19.
Methods
Study design and participants
SPECTRA is an ongoing, double-blind, randomised, placebo-controlled phase 2 and 3 study being done in 31 centres (clinical vaccination centres) in five countries (Belgium, Brazil, Colombia, Philippines, and South Africa). Healthy volunteers (who were monitored for signs of COVID-19) were recruited by the investigators using local advertising. In this report we present the analysis of data obtained between March 24, 2021 (when the first participant was enrolled), and the predefined interim cutoff on Aug 10, 2021, when 150 eligible cases of COVID-19 had been detected to allow the assessment of the primary efficacy objective in adults. For this analysis, eligible participants were male or female adults aged 18 years and older who were in good health or with a stable chronic health condition. An adolescent cohort (aged 12–18 years) was a late addition to the protocol and results from that group will be reported once completed. The main exclusion criteria were pregnancy, receipt of any ongoing immunosuppressive therapy, a history of anaphylaxis to any vaccine component, or previous receipt of any other COVID-19 vaccine. Women with childbearing potential were required to use an approved form of contraception from 30 days before their first dose until 90 days after their second dose. Men were required to use an approved form of contraception from the day of their first dose until 6 months after their second dose, and required not to donate sperm during this period. Detailed inclusion and exclusion criteria are provided in the appendix (pp 2–3).
The study was designed with the collaboration of the Coalition for Epidemic Preparedness Innovations, who provided funding for the vaccine development, and the study procedures and efficacy criteria were agreed with regulatory authorities (European Medicines Agency, Medicines and Healthcare products Regulatory Agency of the UK, China Center for Drug Evaluation, Anvisa Brazil, and Philippines Food and Drug Administration) before initiation. The protocol was approved by all site institutional review boards and applicable national authorities and the study done in accordance with principles of the Declaration of Helsinki and the Good Clinical Practice guidelines of the International Council for Harmonisation. Oversight was provided by an independent unmasked data safety monitoring board who regularly reviewed all safety data, and all cases were adjudicated by a masked independent expert committee. All participants provided written informed consent at enrolment.
Randomisation and masking
Random assignment was stratified by age (cohorts 18–64 years and ≥65 years), the absence or presence of comorbidities associated with a high risk of severe COVID-19, and a known history of COVID-19. The Cenduit Interactive Response Technology system (IQVIA, Durham, NC, USA) was used to randomly assign participants (1:1), using a block size of six, to receive two doses of either SCB-2019 or placebo, with 21 days between doses. Blocks were dynamically assigned to each site for each stratum from a central block pool on the first participant enrolment into the stratum. Subsequent participants enrolled into the site strata were allocated to the next available treatment group in the randomisation block. The randomisation lists were generated by external unmasked statisticians who played no further role in endpoint analyses. All other study staff and the participants were masked to group assignment.
Procedures
Data was collected on sex, age, risk of severe COVID-19 (measured through the presence of known comorbidities associated with COVID-19 risk), ethnicity, race, body-mass index, SARS-CoV-2 status, history of COVID-19, and country at baseline. Although the final commercial formulation of this vaccine will be presented in two vials (one for the vaccine and one for CpG-1018 for mixing pre-administration), for this experimental study the vaccine was supplied in three containers: a prefilled syringe containing 720 μg SCB-2019 in 1·0 mL phosphate-buffered saline, the CpG-1018 adjuvant (Dynavax Technologies, Emeryville, CA, USA) in a 2·0 mL vial containing 12 mg/mL of a 22-mer phosphorothioate oligodeoxynucleotide in Tris buffered saline (24 mg per vial), and alum in vials of 10 mg/mL aluminium hydroxide (Alhydrogel, Croda Health Care). All components were stored in refrigerators at 2–8°C. Unmasked vaccine administrators who did not take part in any other aspect of the study mixed the components according to the pharmacy manual, so the final vaccine formulation contained 30 μg SCB-2019 adjuvanted with 1·50 mg CpG-1018 and 0·75 mg alum per dose. A serum sample was first obtained from the participants to establish their serostatus with respect to SARS-CoV-2 using an ELISA S protein test for the later stratification of the analysis to seronegatives as per protocol. The administrator then gave a 0·5 mL dose of the vaccine or placebo (0·9% sodium chloride for injection supplied in 10 mL ampoules from local manufacturers) by intramuscular injection in the deltoid of the non-dominant arm. Participants were monitored for 30 min after each injection.
Nasal pharyngeal swabs were collected in viral transport medium tubes and RNA extracted. Purified RNA was reverse transcribed into cDNA and used for library preparation with the Illumina COVIDseq protocol (Illumina, San Diego, CA, USA). Briefly, synthesised cDNA was PCR amplified with SARS-CoV-2 specific primer pools to generate 98 amplicons across the SARS-CoV-2 genome (ARTIC multiplex PCR, Eurofins Genomics, Louisville, KY, USA). The primer pools also contained primers targeting human RNA to generate 11 amplicons as controls. PCR amplified fragments were processed for adaptor ligation, enrichment, and clean up. Pooled fragments were quantified, and the fragment sizes were analysed to normalise the amplicon to adaptor concentration. For sequencing, pooled libraries were denatured, neutralised, and loaded onto the Illumina workflow to carry out Illumina sequencing. Raw data generated from the Illumina workflow was processed using the DRAGEN COVIDseq test for a quality check and sequence assembly reporting consensus SARS-CoV-2 sequence. Pangolin and NextClade were used for viral variants and lineages identification.
Outcomes
There were two coprimary objectives with corresponding endpoints: the reactogenicity of the vaccine in an embedded phase 2 study, and the efficacy of SCB-2019 against COVID-19 in participants with no previous exposure to SARS-CoV-2 in the phase 3 study.
The primary reactogenicity endpoint was based on an embedded phase 2 study planned for the first 1600 participants enrolled, 800 each in the vaccine and placebo groups. Participants in this cohort completed electronic diaries (ePRO; Castor, Hoboken, NJ, USA) soliciting local reactions and systemic adverse events for 7 days after each injection, and any unsolicited adverse events up to study day 43 (14 days after the second dose). Data are presented as percentages of each study group with an event according to the highest severity. Safety is being assessed in an ongoing safety follow-up planned for 12 months after the second vaccination in the safety set that includes any participant who received at least one dose of vaccine or placebo. All participants in the safety set were to notify their study centre immediately in the event of any serious adverse event, adverse event of special interest, or any medically attended adverse event throughout the study.
The primary efficacy analysis was based on COVID-19 data obtained up to the cutoff date of Aug 10, 2021, in the per-protocol population whose eligible participants were those with no major protocol deviation that could affect the results of the efficacy analysis and were seronegative for SAR-CoV-2 S protein at baseline with no medical history of COVID-19.16 Efficacy was also estimated for the full analysis set, which consisted of all those who received both injections and provided data for the efficacy analysis, irrespective of their baseline serostatus. Procedures for establishing efficacy are described in the appendix (p 3). Briefly, COVID-19 cases were identified in two ways. Participants used the ePRO system to spontaneously report prespecified symptoms or the study team assessed suspected symptoms at one of the once per week contacts (appendix p 4). Additionally, all participants were supplied with Rapid COVID-19 Antigen Testing kits (Roche Diagnostics, Basle, Switzerland) for once per week self-testing after extensive training at initial site visits. Any positive test with or without symptoms was reported to the study centre for further testing. When symptoms were verified or Rapid COVID-19 Antigen Testing was positive, a nasopharyngeal swab was collected within 2–5 days of onset for RT-PCR confirmation of SARS-CoV-2. Participants with confirmed COVID-19 were monitored daily for 10 days or until case resolution, with a daily recording of body temperature, heart rate, and oxygen saturation by pulse oximetry. The investigators assessed the severity of any COVID-19 case 4 weeks after onset according to the definitions in the appendix (pp 5–6). The primary and secondary efficacy endpoints of each case were adjudicated for consistency and compliance with case definitions by an independent endpoint adjudication committee composed of independent experts who were masked to study group assignment of each case.
The key secondary outcomes we examined, which had been prespecified a priori in the statistical analysis plan and are reported here, include efficacy in the per-protocol population against moderate-to-severe and severe COVID-19 and COVID-19-associated admission to hospital, and vaccine efficacy according to identified virus lineage in the per-protocol population. We also analysed efficacy against all and different severities of COVID-19 in the full analysis set, comprising all participants who received their injections on schedule with no major protocol deviations.
Statistical analysis
The study sample size was driven by the primary efficacy objective. The target for final analysis was 150 eligible cases of any RT-PCR-confirmed COVID-19 in the per-protocol population, which would provide approximately 90% power to reject the null hypothesis (vaccine efficacy ≤30% for COVID-19 with any severity), assuming the true vaccine efficacy was at least 60%. With an attack rate for any COVID-19 strain of 0·60% per month in the placebo group, and participants followed up for approximately 2·04 months for the primary efficacy endpoint, 30 174 participants were enrolled, assuming the non-evaluability was 40% or less.
The primary efficacy endpoint was the first occurrence of RT-PCR-confirmed COVID-19 of any severity, with onset at least 14 days after the second vaccination (the point at which the peak immune response to the vaccine was expected to be achieved) in the per-protocol population consisting of participants who were seronegative for SARS-CoV-2 at baseline and received all study injections on schedule in accordance with US Food and Drug Administration (FDA) guidance.16 For the primary endpoint, the null (H10) hypothesis was that vaccine efficacy is equal to or less than 30%, and alternative (H1a) hypothesis was that vaccine efficacy is more than 30%. Vaccine efficacy is calculated as 100 × (1–incidence rate ratio). The incidence rate is the number of participants with any RT-PCR-confirmed COVID-19 of any severity divided by cumulative follow-up person-time among all participants at risk. Vaccine efficacy in the final analysis is shown if the lower limit of the adjusted CI for vaccine efficacy against COVID-19 of any severity exceeds 30%. This primary objective is evaluated on the basis of the exact binomial method and type 1 adjusted CIs.
One interim efficacy analysis was planned to be conducted only when 50% of the target events (75 cases of RT-PCR confirmed COVID-19) had been reported across the active and control groups. For the interim analysis, the gamma (−2) spending function was used for efficacy boundary specification. This interim analysis evaluated the primary efficacy objective only. Vaccine efficacy was to be declared if the primary objective was met at the interim; otherwise, the study was to be continued. This interim analysis used an alpha of 0·0067 with a type 1 adjusted CI of 98·66%. For the final analysis with 150 cases we used an α of 0·0214 with a type 1 adjusted CI of 95·72%. With a type 1 error of 0·0067 for the interim analysis, and 0·0214 for the final efficacy analysis, an overall type 1 error is maintained at 0·025 (one-sided).
Vaccine efficacy was reported as a relative risk reduction and the absolute risk reduction, which is the absolute difference of attack rates (the percentage of an at-risk population that contracts the disease during a specified time interval) with and without a vaccine, for which the 95% CI was calculated by the Clopper-Pearson method.17 Data are presented as Kaplan-Meier plots of COVID-19 incidence in at-risk populations in the per-protocol cohort and the full analysis set. Vaccine efficacy was also expressed as the number needed to vaccinate to prevent one case of COVID-19, which is the reciprocal of the absolute risk reduction (1/absolute risk reduction).
Efficacy in the per-protocol population against moderate-to-severe and severe COVID-19 and COVID-19-associated admission to hospital were evaluated with type 1-adjusted CIs. Vaccine efficacy according to identified virus lineage was evaluated with 95% CI as was efficacy against all and different severities of COVID-19 in the full analysis set. In accordance with US FDA recommendations,16 the predefined criterion for showing vaccine efficacy in secondary analyses was if the lower limit of the adjusted CI for the vaccine efficacy was greater than 0%. Statistical analyses were done using SAS 9.4.
An independent data and safety monitoring board was convened to review the cumulative study data to evaluate the safety, study conduct, scientific validity, and data integrity of the study to assess its progress and provide recommendations to the sponsor during the entire study period.
This study is registered on EudraCT (2020–004272–17) and ClinicalTrials.gov (NCT04672395).
Role of the funding source
Authors who are employees of Clover Biopharma-ceuticals (IS, HHH, PLi, JL, and BH) or scientific advisers for the study (FR, RC, DA, PR, and GS) participated in the design and development of the protocol, data analysis, and interpretation. The funder (the Coalition for Epidemic Preparedness Innovations) reviewed the protocol. IS, HHH, PLi, FR, and RC worked with a medical writer financed by Clover Biopharmaceuticals to prepare a first draft manuscript that was reviewed and revised by all authors to create the final draft.
Results
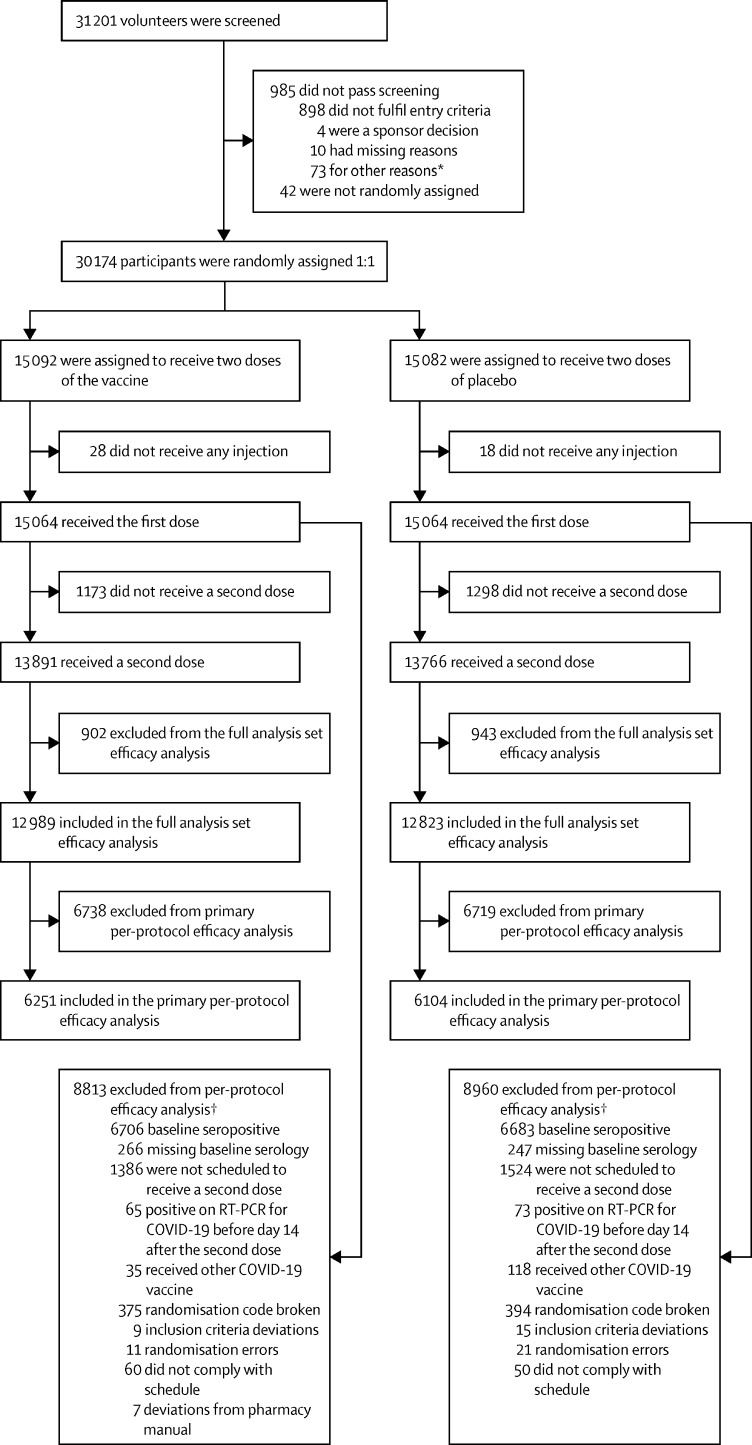
Of 31 201 screened volunteers, 30 174 were enrolled and randomly assigned to a group from March 24 to July 19, 2021 (figure 1 ). A total of 30 155 had valid baseline serological data, of whom 13 389 (44·4%) were seropositive; seropositivity rates for the different countries were 11·3% (80 of 709) in Belgium, 25·0% (1992 of 7974) in Brazil, 36·7% (2456 of 6696) in Colombia, 61·5% (8406 of 13 676) in the Philippines, and 41·4% (455 of 1100) in South Africa. In total 30 128 received a first dose of SCB-2019 (15 064 participants) or placebo (15 064 participants); all these participants constituted the safety set (table 1 ). After exclusions from this group, the full analysis set consisted of 25 812 participants who received both doses (12 989 in the vaccine group and 12 823 in the placebo group) and were analysed for vaccine efficacy. Of these, 6251 in the vaccine group and 6104 in the placebo group were baseline seronegative individuals who were included in the per-protocol population efficacy analyses. The mean age in the safety set was 32·1 years, ranging from 18 to 86 years, and 18·1% (5463 of 30 128) had known underlying comorbidities, putting them at an increased risk of severe COVID-19 disease according to US FDA guidance.16
Figure 1.
Study flow chart
*Other reasons included administrative reasons, contraception requirements being unacceptable, personal medical reasons, withdrawal by volunteers before receiving dose one, etc. †Participants could have more than one reason for being excluded from per-protocol analysis.
Table 1.
Demographics at baseline of the randomly assigned, exposed population (safety set) and the per-protocol population
|
Safety set |
Per-protocol population |
|||||
|---|---|---|---|---|---|---|
| Total (n=30 128) | SCB-2019 (n=15 064) | Placebo (n=15 064) | SCB-2019 (n=6251) | Placebo (n=6104) | ||
| Sex | ||||||
| Male | 16 009 (53·1%) | 7978 (53·0%) | 8031 (53·3%) | 3392 (54·3%) | 3391 (55·6%) | |
| Female | 14 119 (46·9%) | 7086 (47·0%) | 7033 (46·7%) | 2859 (45·7%) | 2713 (44·4%) | |
| Mean age, years | 32·1 (18–86) | 32·1 (18–86) | 32·0 (18–81) | 31·2 (18–79) | 31·0 (18–80) | |
| Age group | ||||||
| ≥18 to 64 | 29 712 (98·6%) | 14 863 (98·7%) | 14 849 (98·6%) | 6197 (99·1%) | 6040 (99·0%) | |
| ≥65 to 74 | 366 (1·2%) | 176 (1·2%) | 190 (1·3%) | 49 (0·8%) | 55 (0·9%) | |
| ≥75 | 50 (0·2%) | 25 (0·2%) | 25 (0·2%) | 5 (0·1%) | 9 (0·1%) | |
| At a high risk of severe COVID-19* | 5463 (18·1%) | 2769 (18·4%) | 2694 (17·9%) | 1068 (17·1%) | 992 (16·3%) | |
| Hispanic or Latino ethnicity | ||||||
| Hispanic or Latino | 13 726 (45·6%) | 6857 (45·5%) | 6869 (45·6%) | 3356 (53·7%) | 3224 (52·8%) | |
| Not Hispanic or Latino | 15 875 (52·7%) | 7950 (52·8%) | 7925 (52·6%) | 2767 (44·3%) | 2759 (45·2%) | |
| Not reported and unknown | 527 (1·7%) | 257 (1·7%) | 270 (1·8%) | 128 (2·0%) | 121 (2·0%) | |
| Race | ||||||
| American Indian† or Alaskan Native | 6544 (21·7%) | 3274 (21·7%) | 3270 (21·7%) | 1294 (20·7%) | 1252 (20·5%) | |
| Asian | 13 720 (45·5%) | 6852 (45·5%) | 6868 (45·6%) | 2230 (35·7%) | 2200 (36·0%) | |
| Black or African American | 2979 (9·9%) | 1519 (10·1%) | 1460 (9·7%) | 716 (11·5%) | 705 (11·5%) | |
| Native Hawaiian or Pacific Islander | 7 (<0·1%) | 4 (<0·1%) | 3 (<0·1%) | 2 (<0·1%) | 2 (<0·1%) | |
| White | 6098 (20·2%) | 3022 (20·1%) | 3076 (20·4%) | 1829 (29·3%) | 1759 (28·8%) | |
| Other | 176 (0·6%) | 91 (0·6%) | 85 (0·6%) | 46 (0·7%) | 47 (0·8%) | |
| Unknown or not reported | 604 (2·0%) | 302 (2·0%) | 302 (2·0%) | 134 (2·1%) | 139 (2·3%) | |
| Baseline SARS-CoV-2 status | ||||||
| Negative | 14 993 (49·8%) | 7483 (49·7%) | 7510 (49·9%) | 6251 (100%) | 6104 (100%) | |
| Positive | 14 622 (48·5%) | 7315 (48·6%) | 7307 (48·5%) | 0 | 0 | |
| Missing data | 513 (1·7%) | 266 (1·8%) | 247 (1·6%) | 0 | 0 | |
| Known history of COVID-19 at baseline | ||||||
| No | 28 522 (94·7%) | 14 259 (94·7%) | 14 263 (94·7%) | 6251 (100·0%) | 6104 (100·0%) | |
| Yes | 1602 (5·3%) | 802 (5·3%) | 800 (5·3%) | 0 | 0 | |
| Missing | 4 (<0·1%) | 3 (<0·1%) | 1 (<0·1%) | 0 | 0 | |
| Country | ||||||
| Belgium (3 sites) | 709 (2·4%) | 354 (2·3%) | 355 (2·4%) | 255 (4·1%) | 277 (4·5%) | |
| Brazil (5 sites) | 7947 (26·4%) | 3973 (26·4%) | 3974 (26·4%) | 2258 (36·1%) | 2153 (35·3%) | |
| Colombia (9 sites) | 6696 (22·2%) | 3348 (22·2%) | 3348 (22·2%) | 1330 (21·3%) | 1294 (21·2%) | |
| Philippines (10 sites) | 13 676 (45·4%) | 6834 (45·4%) | 6842 (45·4%) | 2218 (35·5%) | 2177 (35·7%) | |
| South Africa (4 sites) | 1100 (3·7%) | 555 (3·7%) | 545 (3·6%) | 190 (3·0%) | 203 (3·3%) | |
Data shown as number of participants (%) or mean (range).
Risk because of the presence of known comorbidities, including: asthma, cancer, chronic kidney disease, chronic artery disease, cardiomyopathy, type 1 or 2 diabetes, heart failure, chronic obstructive pulmonary disease, hypertension or high blood pressure, immunocompromised, liver disease, obesity with a body-mass index of 30 kg/m2 or more, sickle cell disease, living with HIV, and substance abuse disorders.
Refers to Indigenous peoples of Colombia.
The adjudication of case files obtained before the Aug 10, 2021, cutoff identified 248 RT-PCR-confirmed COVID-19 cases occurring at least 14 days after the second dose in the full analysis set population, and 207 RT-PCR-confirmed COVID-19 cases in the per-protocol population (table 2 ). Viral samples from 213 (86%) cases in the full analysis set and 179 (86%) of the cases in the per-protocol population were sequenced and lineage identified in 169 (68%) cases in the full analysis set and 146 (71%) in the per-protocol population. None of the identified viruses was the original WH-Human1 strain SARS-CoV-2; all identified lineages from full analysis set participants were variants, most being delta (73 cases [43% of identified variants]), mu (38 cases [22%]), and gamma (13 cases [8%]; table 2). Other less prevalent identified variants included alpha, B.1.623, beta, lambda, and theta. A similar distribution was observed in the per-protocol cases (table 3 ).
Table 2.
Variables in full analysis set and per-protocol population for the calculation of vaccine efficacy endpoints
|
Full analysis set |
Per-protocol population* |
|||||
|---|---|---|---|---|---|---|
| Total | SCB-2019 | Placebo | Total | SCB-2019 | Placebo | |
| RT-PCR-confirmed COVID-19 cases | ||||||
| Total adjudicated cases | 248 (100%) | 63 (100%) | 185 (100%) | 207 (100%) | 52 (100%) | 155 (100%) |
| Virus sequenced | 213 (86%) | 54 (86%) | 159 (86%) | 179 (86%) | 45 (87%) | 134 (86%) |
| Virus sequenced and lineage identified | 169 (68%) | 38 (60%) | 131 (71%) | 146 (71%) | 35 (67%) | 111 (72%) |
| Virus sequenced but no lineage identified | 44 (18%) | 16 (25%) | 28 (15%) | 33 (16%) | 10 (19%) | 23 (15%) |
| Virus not yet sequenced† | 35 (14%) | 9 (14%) | 26 (14%) | 28 (14%) | 7 (13%) | 21 (14%) |
| Identified variants | ||||||
| Total identified variants | 169 (100%) | NA | NA | 146 (100%) | NA | NA |
| Delta (B.1.617.2) | 73 (43%) | 13 (8%) | 60 (36%) | 56 (38%) | 10 (7%) | 46 (32%) |
| Gamma (P.1) | 13 (8%) | 1 (1%) | 12 (7%) | 13 (9%) | 1 (1%) | 12 (8%) |
| Mu (B.1.621) | 38 (22%) | 11 (7%) | 27 (16%) | 37 (25%) | 11 (8%) | 26 (18%) |
| Other (alpha, beta, B.1.623, lambda, theta, etc) | 45 (27%) | 13 (8%) | 32 (19%) | 40 (27%) | 13 (9%) | 27 (18%) |
| Country distribution of cases | ||||||
| Total | 248 (100%) | 63 (100%) | 185 (100%) | 207 (100%) | 52 (100%) | 155 (100%) |
| Belgium | 2 (1%) | 0 (<1%) | 2 (1%) | 2 (1%) | 0 (<1%) | 2 (1%) |
| Brazil | 19 (8%) | 6 (10%) | 13 (7%) | 18 (9%) | 5 (10%) | 13 (8%) |
| Colombia | 76 (31%) | 21 (33%) | 55 (30%) | 70 (34%) | 19 (37%) | 51 (33%) |
| Philippines | 136 (55%) | 32 (51%) | 104 (56%) | 102 (49%) | 24 (46%) | 78 (50%) |
| South Africa | 15 (6%) | 4 (6%) | 11 (6%) | 15 (7%) | 4 (8%) | 11 (7%) |
Data shown as number of participants (%). NA=not available.
Per-protocol analysis includes cases of RT-PCR-confirmed COVID-19 of any severity in participants with no evidence of SARS-CoV-2 at baseline with the onset of the virus 14 days or more after the second dose.
Strains not sequenced at the time of this analysis.
Table 3.
Primary and key secondary vaccine efficacy endpoints in per-protocol population
|
SCB-2019 (n=6251) |
Placebo (n=6104) |
Vaccine efficacy rate (CI)* | ||||||
|---|---|---|---|---|---|---|---|---|
| Number at risk | Cumulative follow-up in person-years† | Number with event | Number at risk | Cumulative follow-up in person-years† | Number with event | |||
| Vaccine efficacy in SARS-CoV-2-naive participants | ||||||||
| Any severity RT-PCR-confirmed COVID-19 | 5935 | 517·3 | 52 | 5806 | 506·1 | 155 | 67·2% (95·72% CI 54·3 to 76·8) | |
| Moderate-to-severe RT-PCR-confirmed COVID-19 | 5935 | 517·3 | 6 | 5806 | 506·1 | 36 | 83·7% (97·86% CI 55·9 to 95·4) | |
| Severe RT-PCR-confirmed COVID-19 | 5935 | 517·3 | 0 | 5806 | 506·1 | 8‡ | 100% (97·86% CI 25·3 to 100·0) | |
| Any severity RT-PCR-confirmed COVID-19 associated with admission to hospital | 5935 | 517·3 | 0 | 5806 | 506·1 | 8‡ | 100% (42·7 to 100·0) | |
| Vaccine efficacy endpoints in SARS-CoV-2-naive participants with any severity COVID-19 against specific variants§ | ||||||||
| Delta variant (B.1.617.2) | 5935 | 517·3 | 10 | 5806 | 506·1 | 46 | 78·7% (57·3 to 90·4) | |
| Gamma variant (P.1) | 5935 | 517·3 | 1 | 5806 | 506·1 | 12 | 91·8% (44·9 to 99·8) | |
| Mu variant (B.1.621) | 5935 | 517·3 | 11 | 5806 | 506·1 | 26 | 58·6% (13·3 to 81·5) | |
| Other variants or not identified¶ | 5935 | 517·3 | 13 | 5806 | 506·1 | 27 | 55·0% (24·9 to 73·8) | |
| Vaccine efficacy in SARS-CoV-2-naive participants at low risk and high risk because of absence or presence of underlying comorbidities | ||||||||
| Any severity RT-PCR-confirmed COVID-19 | ||||||||
| Low | 4908 | 427·8 | 38 | 4857 | 423·3 | 117 | 67·9% (53·3 to 78·3) | |
| High | 1027 | 89·5 | 14 | 949 | 82·8 | 38 | 65·9% (35·7 to 82·9) | |
| Moderate-to-severe RT-PCR-confirmed COVID-19 | ||||||||
| Low | 4908 | 427·8 | 3 | 4857 | 423·3 | 23 | 87·1% (57·3 to 97·5) | |
| High | 1027 | 89·5 | 3 | 949 | 82·8 | 13 | 78·7% (22·3 to 96·1) | |
| Severe RT-PCR-confirmed COVID-19 | ||||||||
| Low | 4908 | 427·8 | 0 | 4857 | 423·3 | 1 | 100% (−3759 to 100·0) | |
| High | 1027 | 89·5 | 0 | 949 | 82·8 | 7 | 100% (35·8 to 100·0) | |
| Any severity RT-PCR-confirmed COVID-19 associated with admission to hospital | ||||||||
| Low | 4908 | 427·8 | 0 | 4857 | 423·3 | 2 | 100% (−427 to 100·0) | |
| High | 1027 | 89·5 | 0 | 949 | 82·8 | 6 | 100% (21·5 to 100·0) | |
| Vaccine efficacy in SARS-CoV-2-naive participants with any severity COVID-19 according to age, sex, or BMI | ||||||||
| Age 18–59 years | 5814 | 502·3 | 49 | 5679 | 489·5 | 147 | 67·5% (54·8 to 77·0) | |
| Age ≥60 years | 121 | 15·0 | 3 | 127 | 16·7 | 8 | 58·4% (−73·4 to 92·9) | |
| Female | 2703 | 229·2 | 25 | 2569 | 217·0 | 72 | 67·1% (47·5 to 80·0) | |
| Male | 3232 | 288·1 | 27 | 3237 | 289·1 | 83 | 67·4% (49·1 to 79·7) | |
| BMI <30 | 5054 | 444·0 | 42 | 4987 | 438·8 | 128 | 67·6% (53·7 to 77·7) | |
| BMI ≥30 | 880 | 73·3 | 10 | 817 | 67·2 | 27 | 66·0% (27·6 to 85·3) | |
BMI=body mass index.
CI for vaccine efficacy, calculated using the Clopper-Pearson method based on conditional binomial distribution, was 95% CI unless shown otherwise.
Cumulative follow-up calculated among all participants at risk within each group, using the time period from 14 days after the second dose to the analysis cutoff on Aug 10, 2021.
Of eight severe cases of COVID-19, seven were admitted to hospital; of eight participants admitted to hospital, seven had severe COVID-19 and one had moderate COVID-19 with pneumonia.
Only calculated for variants for which a sufficient number of cases were detected to provide meaningful analysis.
Includes cases where variant was not identified or cases were too few for variant-specific analysis (alpha, beta, B.1.623, lambda, theta, etc).
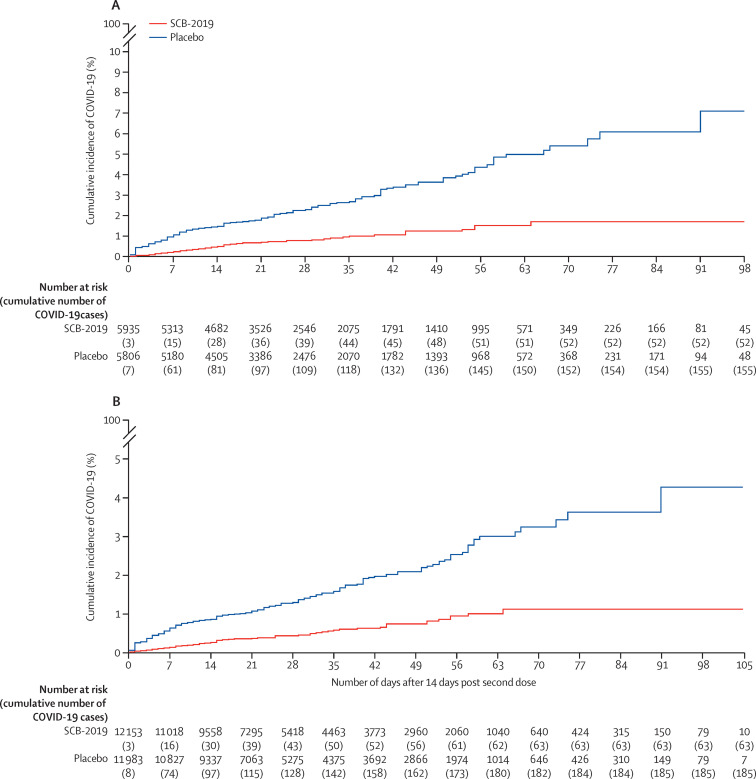
The primary objective assessed in the 207 adjudicated cases in the baseline SARS-CoV-2 seronegative per-protocol population, 52 in the vaccine group (attack rate 0·88%) and 155 in the placebo group (attack rate 2·67%), was a vaccine efficacy of 67·2% (95·72% CI 54·3–76·8) against any severity of RT-PCR-confirmed COVID-19 disease (figure 2A ; table 3). The absolute risk reduction—namely, the difference between attack rates with and without a vaccine—was 1·79% (95% CI 1·31–2·30). Consequently, the number needed to vaccinate to prevent one more case of COVID-19 was 56 (95% CI 44–77). In the per-protocol population there were six cases of moderate-to-severe disease in vaccinees and 36 in placebo recipients, giving a vaccine efficacy of 83·7% (97·86% CI 55·9–95·4). There were no cases of severe COVID-19 in vaccinees versus eight cases in the placebo group, showing a vaccine efficacy of 100% (97·86% CI 25·3–100). Eight participants were admitted to hospital with COVID-19, seven with severe COVID-19 and one with moderate COVID-19 with pneumonia; one participant with severe COVID-19 was not admitted to hospital. All cases of patients being admitted to hospital occurred in placebo recipients, showing a vaccine efficacy against admission to hospital of 100% (95% CI 42·7–100·0). Three COVID-19-related deaths occurred, also all in placebo recipients.
Figure 2.
Kaplan-Meier plots of efficacy of the SCB-2019 plus CpG and alum vaccine candidate against symptomatic COVID-19
Cumulative incidence of RT-PCR-confirmed COVID-19 of any severity with onset at least 14 days after the second dose of SCB-2019 plus CpG and alum or placebo in the per-protocol population with no previous exposure to SARS-CoV-2 (A) and in the full analysis set population (B).
Vaccine efficacy against COVID-19 of any severity due to the three predominant variants individually in the per-protocol population were 78·7% (95% CI 57·3–90·4) for delta, 58·6% (13·3–81·5) for mu, and 91·8% (44·9–99·8) for gamma. For the 40 cases of COVID-19 which were due to other lineages or where the virus could not be identified, of which there were 13 cases in vaccinees and 27 cases in the placebo group, the vaccine efficacy was 55·0% (95% CI 24·9–73·8) against COVID-19 of any severity, and 90·2% (31·2–99·8) against moderate-to-severe disease.
When stratified for their baseline risk of severe COVID-19 due to the presence of known comorbidities, there were 52 cases of any severity of COVID-19 in those at a high risk: 14 of 1027 participants in the vaccine group and 38 of 949 participants in the placebo group (table 3). Vaccine efficacy in this group at a high risk was 65·9% (95% CI 35·7–82·9) against any severity of RT-PCR-confirmed COVID-19 disease and 78·7% (22·3–96·1) against moderate-to-severe disease. There were seven cases of severe disease and six that led to admission to hospital, all of which occurred in the placebo group.
The vaccine efficacy against any severity of COVID-19 was consistently in the range of 66·0–67·6% (table 3) when assessed in the per-protocol population according to age (for those aged 18–59 years), male or female sex, or the generally accepted obesity index (body-mass index ≥30 kg/m2). There were insufficient numbers in the older age group (≥60 years) for a meaningful assessment.
In the full analysis set, including those who were seropositive at baseline, there were 248 cases of confirmed COVID-19 of any severity, with a median follow-up of 74 days. These cases occurred in 63 of 12 153 people at risk (1070·2 person-years of follow-up) in the vaccine group, and 185 of 11 983 people at risk (1045·8 person-years of follow-up) in the placebo group (figure 2B; appendix p 7), giving SCB-2019 a vaccine efficacy of 66·7% (95% CI 55·5–75·4) against any severity of RT-PCR-confirmed COVID-19 disease independent of the baseline serostatus. In the full analysis set, vaccine efficacy against severe COVID-19 or admission to hospital associated with COVID-19 was 100% (42·7–100·0). Against the three predominant variants, the efficacy rates were similar to those in the per-protocol population: 78·8% (61·0–89·3) for delta, 91·9% (45·0–99·8) for gamma, and 60·2% (17·1–82·2) for mu (appendix p 7).
Reactogenicity was assessed in the phase 2 subset of 1601 participants: 808 vaccinees and 793 who received placebo. This assessment showed that the vaccination was generally well tolerated, with 290 (36·1%) of 803 vaccinees and 89 (11·3%) of 786 placebo recipients reporting local adverse reactions after the first dose (appendix p 8). The difference between the groups was because of more reports of mainly mild-to-moderate injection site pain; in 287 (35·7%) of 803 vaccinees versus 81 (10·3%) of 786 placebo recipients (appendix pp 9–10). The rates of solicited local adverse reactions (28·2% in vaccinees and 8·2% in the placebo group) were lower after the second dose, also mainly because of local injection site pain (in 189 [26·9%] of 702 vaccinees vs 52 [7·4%] of 699 placebo recipients). The reported rates of solicited systemic adverse events in vaccine (288 [36%] of 803) and placebo (268 [34%] of 786) groups after the first dose were similar and were generally lower after the second dose (162 [23%] of 702 in the vaccine group and 147 [21%] of 699 in the placebo group). Most solicited local reactions and systemic adverse events were described as mild or moderate, with some transient severe adverse events reported in both vaccine (n=34) and placebo (n=48) groups (appendix p 10).
A safety assessment is ongoing in all 30 128 participants in the phase 3 part of the study who received a first dose of vaccine or placebo (appendix p 9), and an in-depth analysis of safety will be reported separately, including the effect of previous exposure to SARS-CoV-2. The independent data safety monitoring board have not identified any concerns that warranted a pause or modification in the study. To date, the occurrence of unsolicited adverse events has been balanced between the two groups (12·3% in vaccinees vs 12·4% in placebo recipients). Of the 16 deaths reported up to the safety cutoff date, there were three in the vaccine group and 13 in the placebo group, including the three related to COVID-19 disease (appendix p 8). Serious adverse events have been reported by 49 of 15 064 (0·3%) vaccinees and 59 of 15 064 (0·4%) placebo recipients, but vaccine-related cases were rare. Five participants were considered to have treatment-related events: upon unmasking these were found to consist of four vaccinees in whom there were individual cases of moderate hypersensitivity, mild Bell's palsy, spontaneous abortion occurring 31 days after the first vaccination, and an anaphylactic reaction 3 days after the second vaccination. The fifth was a life-threatening case of COVID-19 with respiratory failure and pneumonia 53 days after the second placebo injection.
Discussion
This ongoing study showed a vaccine efficacy of 67·2% (95·72% CI 54·3–76·8) against any severity of RT-PCR-confirmed COVID-19, and 83·7% (97·86% CI 55·9–95·4) efficacy against moderate-to-severe COVID-19 in the per-protocol population of participants without previous exposure to SARS-CoV-2. Efficacy against admission to hospital was 100% and there were no COVID-19-related deaths in the vaccine group, although the low numbers of such cases in the placebo recipients mean these estimates have very wide CIs. Furthermore, the study also showed a vaccine efficacy of 66·7% (95% CI 55·5–75·4) efficacy against any severe COVID-19 and 82·5% (60·3–93·4) against moderate-to-severe COVID-19 in the entire study population independent of baseline serostatus, which represents a real-world scenario. These estimates are consistent with the predicted efficacy based on comparing the S protein binding antibody concentrations after SCB-2019 with those of four authorised vaccines with known efficacy.18 These results were achieved in the context of all cases with identified lineages being due to SARS-CoV-2 variants and an attack rate of 2·67% in the seronegative placebo recipients, which is far greater than in previous efficacy studies of authorised vaccines.7, 8, 9, 10, 11 The number needed to vaccinate of 56 (95% CI 44–77) for SCB-2019 is lower than the range of estimated values of 78–119 for the authorised vaccines mRNA-1273, BNT162b2, ChAdOx1 nCov-19, and Ad26.COV2.s.19 The absolute risk reduction for SCB-2019 was 1·79% (95% CI 1·31–2·30), which is higher than the range of values, 0·84–1·3%, calculated for the aforementioned authorised vaccines.19
Currently used vaccines were authorised on the basis of efficacy estimates established in late 2020,7, 8, 9 when the circulating COVID-19 virus was almost entirely the original WH-Human1 virus, with increasing contributions from alpha and beta variants.20 The epidemiology has now changed with the emergence of new variants: delta (B.1.617.2) in India in September, 2020; gamma (P.1) in Brazil in November, 2020; and mu (B.1.621) in Colombia in January, 2021.21 Clinical efficacy assessments of new SARS-CoV-2 vaccines should take into account this evolution of the pandemic due to the continuing mutation and emergence of variants. This changing epidemiology is clearly illustrated in our study across four continents where no infection with the original WH-Human1 virus was detected. All cases of COVID-19 were due to viral variants, most notably the delta variant, which caused 73 (34%) of 213 cases in which the lineage was identified. This finding is consistent with the evolving epidemiology of SARS-CoV-2 variants; as of Nov 18, 2021, the data suggest that COVID-19 globally is approaching being 100% due to the delta variant.20 Since then a new variant of concern, omicron (B.1.1.529), which was reported initially in South Africa on Nov 24, 2021, has subsequently been identified globally.22 Therefore, the secondary estimates of efficacy against any severity of COVID-19 due to delta variant of 78·7% in the per-protocol population and 78·8% in the full analysis set population might be the most relevant indicator of the probable effectiveness of SCB-2019 if widely used now. This is an important consideration because the delta variant is associated with increased transmission and severity of COVID-19.23 These data are similar to those from approved vaccines; preliminary estimates of effectiveness against symptomatic COVID-19 due to the delta variant indicate that after two doses, the mRNA BNT162b2 vaccine had 88% effectiveness and the vector vaccine, ChAdOx1 nCoV-19, had 67·0%.24 It is also reassuring that SCB-2019 had efficacies of 58·6% (in the per-protocol population) and 60·2% (in the full analysis set) against the recently emerged mu strain (B.1.621), because this new strain has been reported to be resistant to antibodies from convalescent COVID-19 patient serum samples and mRNA BNT162b2 vaccinees.25 Although these vaccine efficacy estimates for the variants were based on low numbers of cases, they were sufficient to have lower CI limits of more than 0%, the prespecified criterion considered to show successful efficacy in these secondary analyses.16 In these circumstances, the efficacy of SCB-2019 in the almost 50% of the study population who were excluded from the primary analysis because of baseline seropositivity from previous SARS-CoV-2 exposure will be equally as important as the per-protocol population. The ongoing analyses of protection afforded by previous infection and the additional efficacy induced by the vaccine in this population will be reported separately.
After the first dose, rates of solicited local (36·1%) and systemic adverse events (35·9%) were lower than those previously reported for the mRNA,7, 8 vector,9, 10 and other subunit vaccines11 for which 49–84% of first doses were associated with solicited local reactions and 46–72% with systemic adverse events. These rates decreased after the second doses, although they have been reported to increase after the second dose of some of the authorised vaccines.8, 11 Initial data suggest that SCB-2019 is safe, with low rates of serious adverse events or medically attended adverse events. Although the surveillance period is relatively short, with a mean of 82 days of follow-up, and needs to be confirmed by detailed analyses from the ongoing surveillance that is intended to continue up to 12 months after the second vaccination, this initial profile is promising. Further planned study will also include an analysis of the immune responses from subsets of participants in the study to investigate the kinetic profile of the immune response and cross-reactivity with the variants.
Limitations of this study were mainly related to factors beyond our control: the rapid spread of the SARS-CoV-2 pandemic with variable rates of asymptomatic infections, the progress of worldwide mass vaccination campaigns starting in older populations, and the changing nature of the infecting SARS-CoV-2 virus. The high rate of SARS-CoV-2 transmission in the study locations resulted in large proportions of the participants being seropositive at baseline such that only 41% (12 355 of 30 128) of the treated population was eligible for the per-protocol analysis. However, the vaccine efficacy estimates for the full analysis set and per-protocol population were similar. Furthermore, the high attack rates and substantial proportion of participants with comorbidities (18%) resulted in a sufficient number of cases with severe disease (0·14%) in seronegative placebo recipients to show substantial efficacy against severe disease. Mass vaccination with authorised vaccines initially targeted older adults, resulting in few unvaccinated older participants being available for the study. However, it is notable that the five COVID-19 cases in participants aged 65 years or older all occurred in placebo recipients. Finally, we have shown clinically meaningful efficacy against the variants that have supplanted the original WH-Human1 virus and were in circulation at the time of our study, most notably the delta variant. The ongoing long-term surveillance will establish the duration of such efficacy and provide a more detailed safety analysis. Another limitation is the requirement for premixing of the vaccine components supplied separately, because the timelines for this clinical development, in the urgency imposed by the COVID-19 pandemic, have not allowed for the manufacturing process to be completed since the clinical decision was made to use this formulation based on the phase 1 study results.14, 15 A clinical study to bridge the immunogenicity data from this experimental formulation with that of the final commercial formulation is anticipated.
This study shows that SCB-2019 plus CpG and alum has approximately 67% efficacy against any severe COVID-19 and 84% efficacy against moderate-to-severe COVID-19, and 100% efficacy against severe disease and admission to hospital because of SARS-CoV-2, including disease due to the predominant variants currently circulating. It has a favourable safety and reactogenicity profile compared with some of the currently available vaccines, and the requirements for storage at normal refrigerator temperatures will greatly facilitate its distribution and use worldwide compared with some of those vaccines that require much lower temperatures for storage.
Data sharing
Once the study is completed the datasets, including the redacted study protocol, redacted statistical analysis plan, and individual participant data supporting the results reported in this Article, will be available 3 months from the initial request to researchers who provide a methodologically sound proposal, at the discretion of the company governing body. The data will be provided after its deidentification, in compliance with applicable privacy laws, data protection, and requirements for consent and anonymisation.
Declaration of interests
IS, HHH, PLi, RH, CB, BH, and JL are full-time employees of Clover Biopharmaceuticals. FR is a statistical adviser for Clover Biopharmaceuticals. RC, DA, PR, and GS are scientific advisers for Clover Biopharmaceuticals. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
The study was funded by Clover Biopharmaceuticals and the Coalition for Epidemic Preparedness Innovations. We thank the participants in the trial and Yung Huang, Lynn Chen, Pilar Rubio, Carole Verhoeven, Haijing Qin, Vincent Mwangi, and Joyce Garcia at Clover Biopharmaceuticals, and our external service providers (contract research organisations, laboratories, clinical suppliers, and biostatistics providers) for their invaluable assistance in the trial. We also thank Dynavax Technologies for providing the CpG 1018 adjuvant. We are grateful to the Scientific Advisory Board members and Coalition for Epidemic Preparedness Innovations team for their advice and guidance, to the members of the data and safety monitoring board for their dedication to the trial (Terry Nolan [Chair], Xiaoping Dong, Catherine Slack, Yola Moride, Scott Evans, Renato Kfouri, Xiaohua Sheng, and Jim Buttery), and to the endpoint adjudication committee (Kathryn B Anderson [Chair], Hans Bock, Kristopher M Paolino, Stephen J Thomas, Olivier Godeaux, Astrid Borkowski, Ricardo Rüttimann, Margarita Riera, Omar Okasha, and Ward Schrooten) for their expert analysis of the trial data. We also thank Keith Veitch (keithveitch communications, Amsterdam, Netherlands) for assistance in writing and preparing the manuscript for submission (funded by Clover Biopharmaceuticals).
Contributors
IS, HHH, PLi, JL, BH, FR, RC, DA, PR, and GS designed the study and prepared the protocol. Data analysis was done by an external service provider supervised by PLi. All authors did the interpretation and writing of the manuscript, led by RC. All authors had access to the analysed datasets. PLi, IS, HHH, and RC had access to and verified the raw data. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supplementary Material
References
- 1.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins University of Medicine COVID-19 Dashboard. https://coronavirus.jhu.edu/map.html
- 3.WHO Draft landscape and tracker of COVID-19 candidate vaccines. Nov 16, 2021. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- 4.Our World in Data Statistics and research: coronavirus (COVID-19) vaccinations. https://ourworldindata.org/covid-vaccinations
- 5.Jara A, Undurraga EA, González C, et al. Effectiveness of an inactivated SARS-CoV-2 vaccine in Chile. N Engl J Med. 2021;385:875–884. doi: 10.1056/NEJMoa2107715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanriover MD, Doğanay HL, Murat Akova M, et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet. 2021;3948:213–222. doi: 10.1016/S0140-6736(21)01429-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falsey AR, Sobieszczyk ME, Hirsch I, et al. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) COVID-19 vaccine. New Engl J Med. 2021;385:2348–2360. doi: 10.1056/NEJMoa2105290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadoff J, Gray G, Vandebosch A, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against COVID-19. N Engl J Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heath PT, Galiza EP, Baxter DN, et al. Safety and efficacy of NVX-CoV2373 COVID-19 vaccine. N Engl J Med. 2021;385:1172–1183. doi: 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J, Su D, Sun Y, et al. Cryo-electron microscopy structure of S-Trimer, a subunit vaccine candidate for COVID-19. J Virol. 2021;95:e00194–e00221. doi: 10.1128/JVI.00194-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang JG, Su D, Song T-Z, et al. S-Trimer, a COVID-19 subunit vaccine candidate, induces protective immunity in nonhuman primates. Nat Commun. 2021;12 doi: 10.1038/s41467-021-21634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richmond P, Hatchuel L, Dong M, et al. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: a phase 1, randomised, double-blind, placebo-controlled trial. Lancet. 2021;397:682–694. doi: 10.1016/S0140-6736(21)00241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richmond P, Hatchuel L, Pacciarini F, et al. Persistence of the immune responses and cross-neutralizing activity with variants of concern following two doses of adjuvanted SCB-2019 COVID-19 vaccine. J Infect Dis. 2021;224:1699–1706. doi: 10.1093/infdis/jiab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Food and Drug Administration Development and licensure of vaccines to prevent COVID-19: guidance for industry. June, 2020. https://www.fda.gov/media/139638/download
- 17.Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat Med. 1998;17:873–890. doi: 10.1002/(sici)1097-0258(19980430)17:8<873::aid-sim779>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 18.Ambrosino D, Han HH, Hu B, et al. Immunogenicity of SCB-2019 coronavirus disease 2019 vaccine compared with 4 approved vaccines. J Infect Dis. 2021 doi: 10.1093/infdis/jiab574. published online Nov 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olliaro P, Torreele E, Vaillant M. COVID-19 vaccine efficacy and effectiveness—the elephant (not) in the room. Lancet Microbe. 2021;2:e279–e280. doi: 10.1016/S2666-5247(21)00069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nextstrain Genomic epidemiology of SARS-CoV-2 with global subsampling. https://nextstrain.org/ncov/open/global
- 21.WHO Tracking SARS-CoV-2 variants. Sept 2, 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 22.WHO Classification of omicron (B.1.1.529): SARS-CoV-2 variant of concern. Nov 26, 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(B.1.1.529)-sars-cov-2-variant-of-concern
- 23.Bernal JL, Andrews N, Gower C, et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong SWX, Chiew CJ, Ang LW, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (alpha), B.1.315 (beta), and B.1.617.2 (delta) Clin Infect Dis. 2021 doi: 10.1093/cid/ciab721. published online Aug 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uriu K, Kimura I, Shirakawa K, et al. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. New Engl J Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Once the study is completed the datasets, including the redacted study protocol, redacted statistical analysis plan, and individual participant data supporting the results reported in this Article, will be available 3 months from the initial request to researchers who provide a methodologically sound proposal, at the discretion of the company governing body. The data will be provided after its deidentification, in compliance with applicable privacy laws, data protection, and requirements for consent and anonymisation.