Extended Data Fig. 10. Mechanistic model of Nrxn1SS4+→Cbln2→GluD1→NMDAR and Nrxn3SS4+→Cbln2→GluD1→AMPAR signalling pathways.
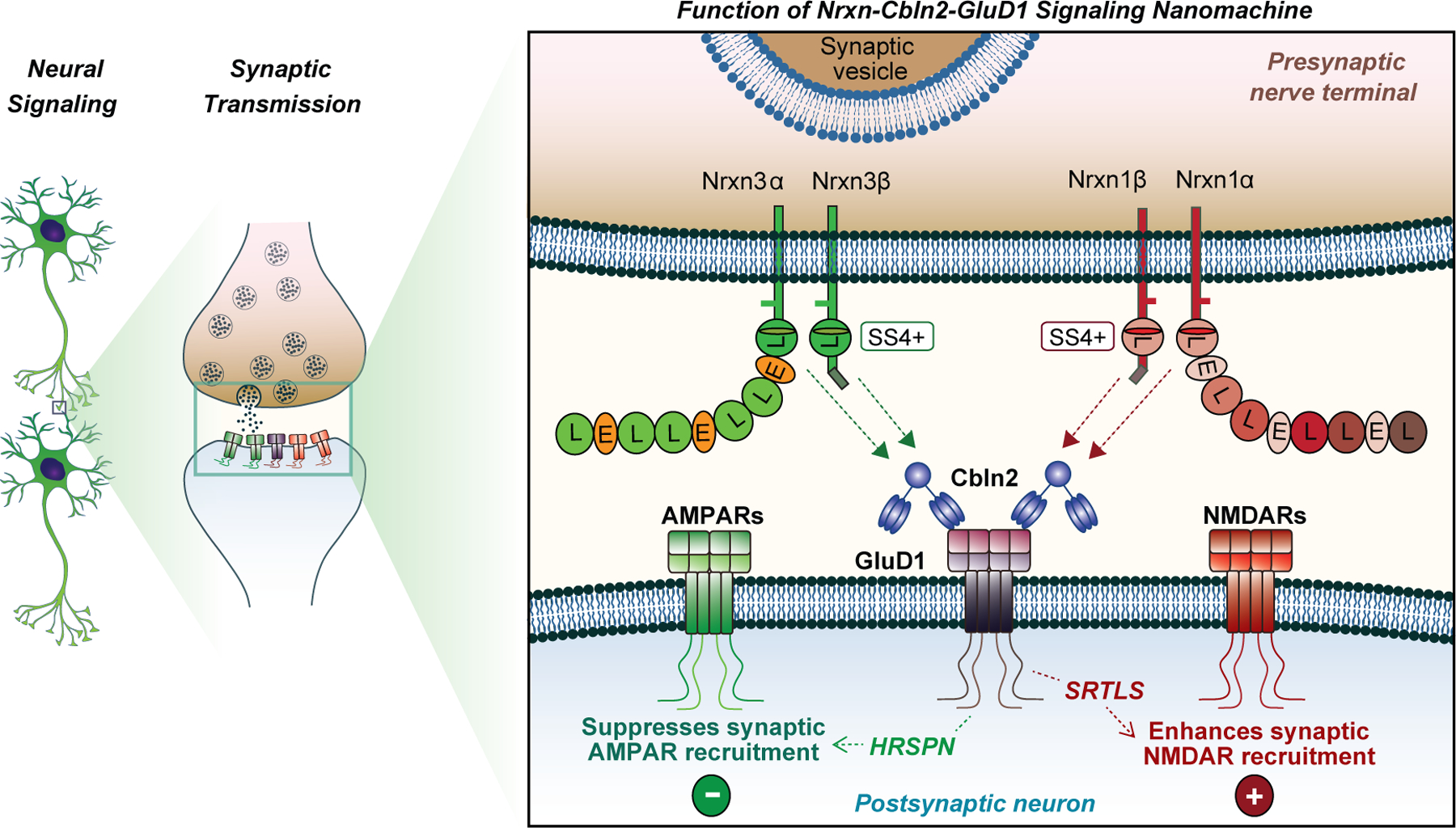
The model, based on the present data, proposes that Nrxn1- and Nrxn3-alternative splicing at SS4 regulates the postsynaptic content of AMPARs and NMDARs at hippocampal synapses in the subiculum but not the CA1 region by binding to Cbln2, which in turn binds to GluD1 (see Figs. 1 and 2). Surprisingly, binding to the same GluD1-Cbln2 complex enables Nrxn1SS4+ to enhance NMDARs but not AMPARs, whereas binding of Nrxn3SS4+ to the GluD1-Cbln2 complex suppresses AMPARs but not NMDARs (see Fig. 3). Signal transduction by the Nrxn1SS4+-Cbln2-GluD1 and Nrxn3SS4+-Cbln2-GluD1 assemblies is mediated by distinct cytoplasmic GluD1 sequences (HRSPN for AMPAR regulation by Nrxn3SS4+, SRTLS for NMDAR regulation by Nrxn1SS4+), suggesting that GluD1 couples Nrxn1SS4+ and Nrxn3SS4+ signals to distinct effector sequences (see Figs. 4 and 5). Since GluD1 and GluD2 constructs lacking the native GluD1 transmembrane architecture are fully active (see Fig. 4 and 5), binding of Nrxn1SS4+-Cbln2 and Nrxn3SS4+-Cbln2 complexes to the ATD of GluD1 (and of GluD2) activates a transmembrane signal without inducing a conformational change in the transmembrane regions of GluDs.
