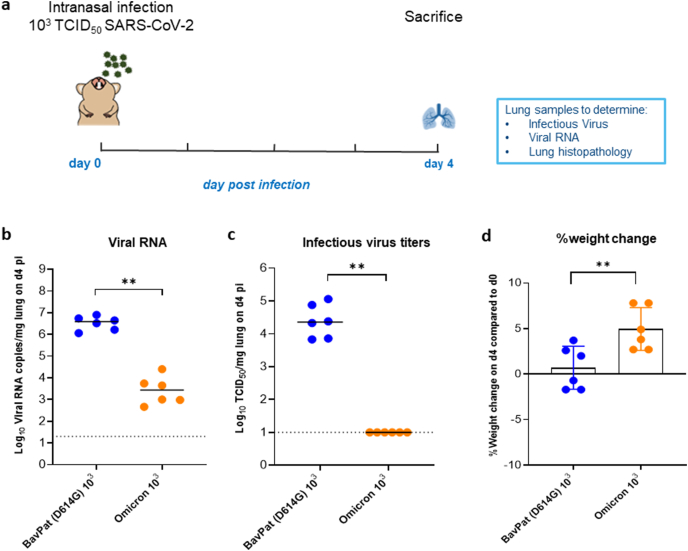
Fig. 1.
Characterization of the in vivo replication of the omicron SARS-CoV-2 variant versus the ancestral D614G strain. (a) Set-up of the Syrian hamster infection study. (b) Viral RNA levels in the lungs of hamsters infected with 103 TCID50 of BavPat (D614G) strain (n = 6) or the omicron (B.1.1.529) SARS-CoV-2 variant (n = 6) on day 4 post-infection (pi) are expressed as log10 SARS-CoV-2 RNA copies per mg lung tissue. Individual data and median values are presented. (c) Infectious viral loads in the lungs of hamsters infected with the D614G strain or the omicron variant at day 4 pi are expressed as log10 TCID50 per mg lung tissue. Individual data and median values are presented. (d) Weight change at day 4 pi in percentage, normalized to the body weight at the time of infection. Bars represent means ± SD. Data were analyzed with the Mann−Whitney U test, **P < 0.01. All data are from a single experiment.