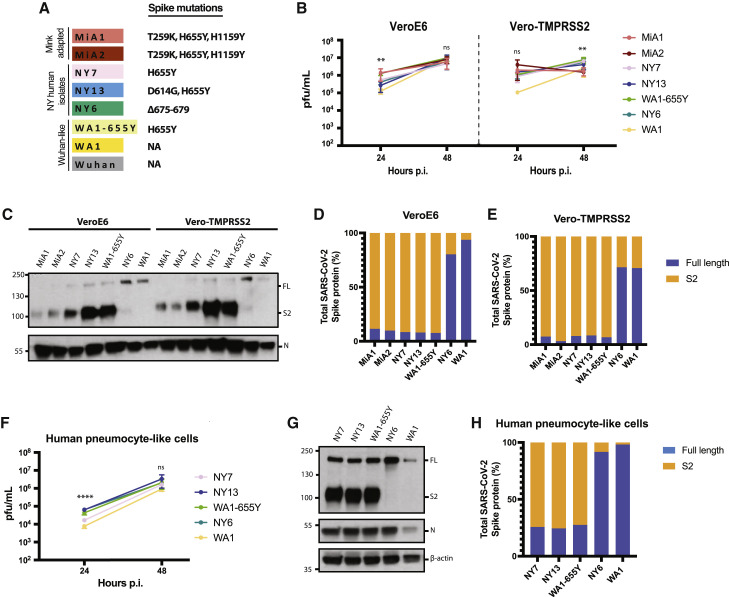
Figure 2.
The H655Y amino acid substitution enhances spike cleavage and viral growth
(A) Spike polymorphisms present in the mink-adapted variants, early human SARS-CoV-2 New York (NY) isolates, WA1-655Y, and WA1 wild-type viruses. Wuhan1 is included as a reference.
(B) Replication kinetics of early SARS-CoV-2 viruses in Vero E6 and Vero-TMPRSS2 cells. Infections were performed at an MOI of 0.01. Viral titers were determined by plaque assay at the indicated hour post-infection and expressed as PFU per milliliter. Shown are the means and SDs from 3 replicates. ANOVA test for multiple comparison was used to compare mean differences within different isolates and time points. Viral isolates were compared two by two using the Tukey’s correction. Statistical significance was considered when p ≤ 0.05 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant). Color codes relate to the isolates shown in (A).
(C) Western blotting of S protein from supernatants of Vero E6- and Vero-TMPRSS2-infected cells. Infections were performed at an MOI of 0.01, and viral supernatants were collected at 48 h post-infection (p.i.). Full-length (FL) spike protein (180 kDa), S2 cleaved spike (95 kDa), and nucleocapsid (N; 50 kDa) were detected using specific antibodies. Levels of N protein were used as loading control.
(D and E) Quantification of full-length and cleaved spike protein of the indicated viruses in Vero E6 and Vero-TMPRSS2 cells. Spike protein levels were normalized to nucleocapsid expression.
(F) Replication kinetics of early SARS-CoV-2 viruses in human pneumocyte-like cells. Cells were infected with 3000 pfu of the corresponding viral isolate per well. Viral titers were determined by plaque assay at the indicated hour p.i. and expressed as PFU per milliliter. Shown are the means and SDs from 3 replicates. ANOVA test for multiple comparison was used to compare mean differences within different isolates and time points. Viral isolates compared two by two using the Tukey’s correction. Statistical significance was considered when p ≤ 0.05 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant).
(G) Western blotting of S protein in human pneumocyte-like cells infected with 3000 pfu of the corresponding early SARS-CoV-2 virus per well. Cell extracts were collected at 48 h p.i. Full-length (FL) spike protein (180 kDa), S2 cleaved spike (95 kDa), nucleocapsid (N; 50 kDa), and β-actin (45 kDa) were detected using specific antibodies. Levels of N and β-actin protein were used as loading control.
(H) Quantification of full-length and cleaved spike protein of the indicated viruses in human pneumocyte-like cells. Spike protein levels were normalized to nucleocapsid expression.