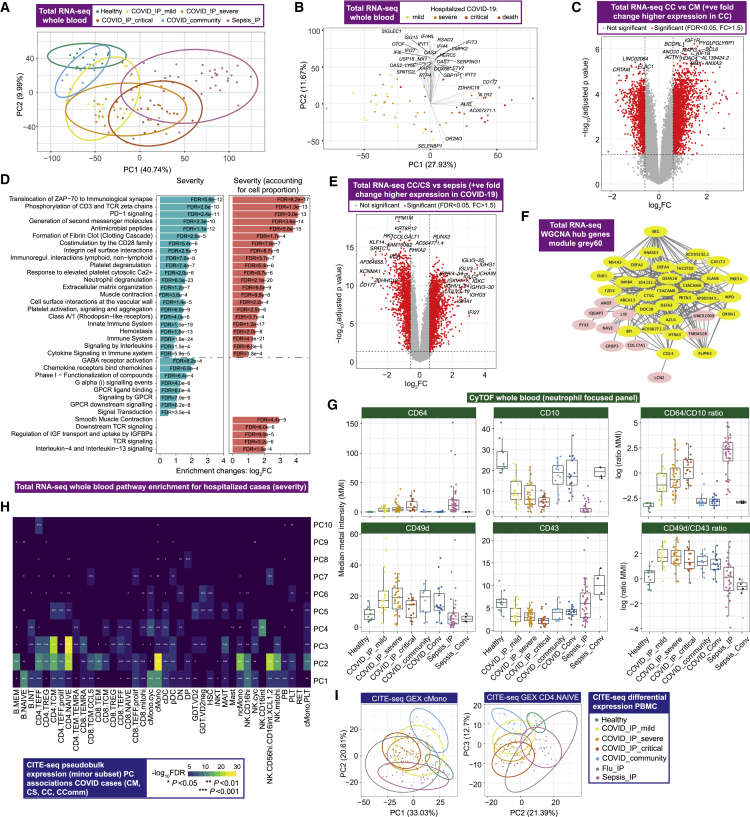
Figure 2.
Signatures of COVID-19 response from transcriptomics
(A–F) Whole blood total RNA-seq.
(A and B) Principal component (PC) analysis of (A) all comparator groups and (B) hospitalized COVID-19 cases.
(C) Differential gene expression critical versus mild COVID-19.
(D) Pathway enrichment for COVID-19 severity as a quantitative trait ± inclusion cell proportion.
(E) Differential gene expression COVID-19 versus sepsis.
(F) Intramodular hub genes for weighted gene correlation network analysis module grey60.
(G) Neutrophil cell surface proteins assayed by mass cytometry shown by marker or ratio of markers. Boxplots show median and first and third quartiles; whiskers show 1.5x interquartile range.
(H and I) CITE-seq gene expression. (H) Association of PCs of expression variance within minor cell subsets in COVID-19 patients. (I) PC plots in classical monocytes and naive CD4+ T cells.
See Figures S3 and S4.