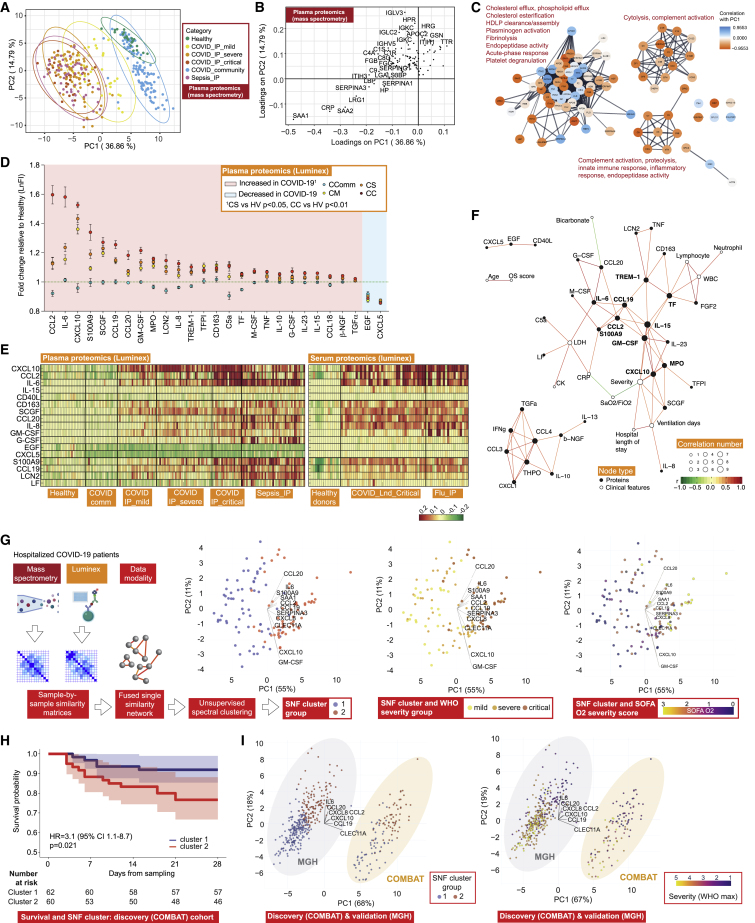
Figure 6.
Plasma protein COVID-19 signatures and sub-phenotypes
(A–C) HT-LC-MS/MS mass spectrometry of plasma proteins.
(A) Principal components analysis (PCA) of all samples.
(B) Proteins contributing to PC loadings (more negative loading values indicating higher positive correlation with disease severity).
(C) Clusters based on protein-protein interaction network with enriched GOBP terms.
(D–F) Proteins significantly differentially expressed between comparator groups assayed by Luminex.
(D) Fold change in plasma proteins in hospitalized COVID-19 versus healthy volunteers. Data represented as mean ± SEM.
(E) Plasma and serum protein abundance by comparator group.
(F) Network of clinical feature−protein correlations in COVID−19 patients and healthy volunteers based on highly correlated events (r2 > 0.7 or < -0.5).
(G) Similarity network fusion (SNF) using plasma proteins for hospitalized COVID-19 patients from COMBAT cohort showing approach and PCA colored by cluster (left) or WHO severity group (middle) or SOFA O2 score (right).
(H) Kaplan-Meier survival plot by SNF cluster group (95% CIs shaded) (HR, hazard ratio calculated using Cox proportional hazard model).
(I) Mass General Hospital (Olink) validation data and COMBAT (discovery) cohorts showing cluster groups (left) or colored by WHO max severity (right).
See Figure S8.