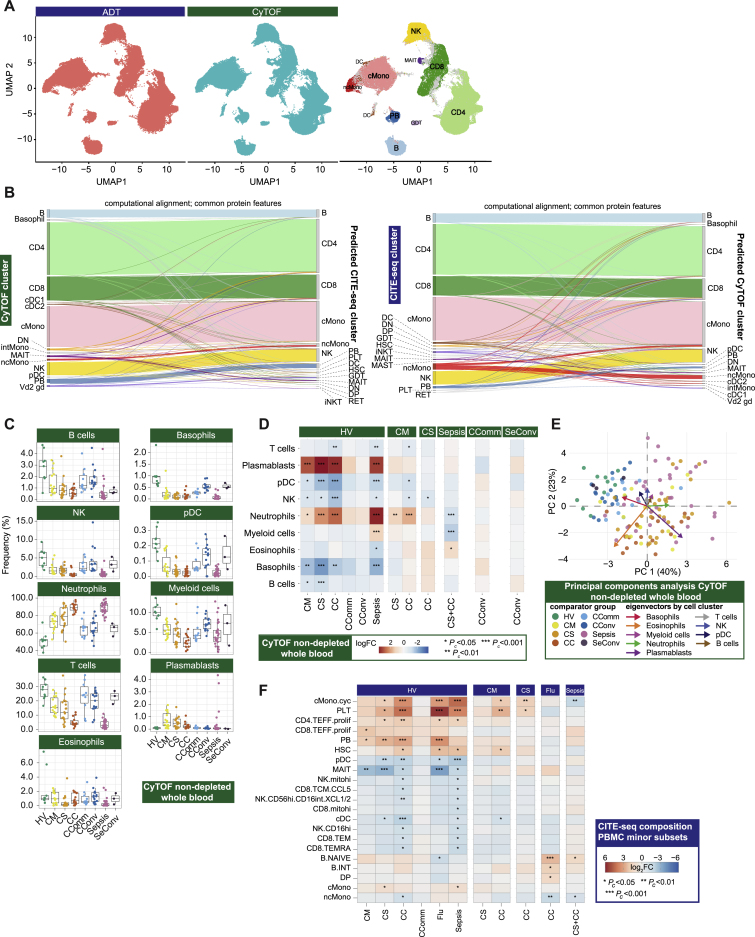
Figure S2.
Single cell compositional approaches, related to Figure 1
(A-B) Concordance and cross validation of cell composition using single cell resolution mass cytometry (Helios CyTOF system) clustering (from granulocyte (CD66+) depleted whole blood with down sampling to a maximum of 75,000 cells and 7,118,158 cells assayed) and CITE-seq analysis of viability sorted peripheral blood mononuclear cells (PBMCs) from 140 samples profiled using the 10X Genomics platform. (A) Concordance in cell composition annotation between assay types is demonstrated with UMAP showing joint visualization of CITE-seq and CyTOF datasets including side by side plot of CITE-seq cell surface protein quantification (ADT) and mass cytometry together with a plot of cell annotations transferred between datasets and colored by cell type where concordant (94.5% of cells) or discordant (gray) (B) Plots demonstrating cross validation of mass cytometry and CITE-seq cell clusters. (C-E) Stabilized whole blood (Cytodelics) from COVID-19 patients (non-granulocyte depleted samples) analyzed by mass cytometry (including matched samples collected during convalescence from 16 COVID-19 hospitalized patients). A self-organizing map algorithm (FlowSOM) resolved 25 clusters by consensus clustering for 3,893,390 cells after down sampling to a maximum of 40,000 cells. Clusters merged to identify broad immune cell populations (Data S3). (C) Cell frequencies by clinical group. Boxplots show median, first and third quartiles; whiskers show 1.5x interquartile range. (D) Differential abundance analysis in patients compared to healthy volunteers, and different disease states clustering major cell populations using empirical Bayes analysis (statistical inference estimating priors from the data). (E) PCA with arrows indicating drivers of variation by cell population. (F) Differential abundance analysis in patients compared to healthy volunteers, and between disease categories for minor cell subsets using empirical Bayes analysis. Abbreviations for CITE-seq (panel F). B: B cell; cDC: classical dendritic cell; cMono: classical monocytes; cyc: cycling; DC: dendritic cell; DN: CD4/CD8 double negative; DP: CD4/CD8: double positive; ERYTH: erythrocyte; GDT: gamma delta T; hi: high; HSC: hematopoietic stem (and progenitor) cells; iNKT: invariant natural killer T; INT/int: intermediate; MAIT: Mucosal associated invariant T; MEM: memory; mito: mitochondrial; MNP: mononuclear phagocyte; ncMono: non-classical monocyte; neg: negative; NK: natural killer cell; PB: plasmablast; PBMC: peripheral blood mononuclear cell; pDC: plasmacytoid dendritic cell; PLT: platelet/CD34- megakaryocyte progenitor; prolif: proliferating; RET: reticulocyte; T: T cell; TCM: T central memory; TEFF: T effector; TEM(RA): T effector memory (CD45RA re-expressing); TREG: T regulatory cell. Comparator group abbreviations. HV: healthy volunteer; CM: COVID-19 in-patient mild; CS: COVID-19 in-patient severe; CC: COVID-19 in-patient critical; CComm: COVID-19 community case in the recovery phase (never admitted to hospital); CConv: COVID-19 convalescence (survivors from 28 days after discharge); Flu: influenza in-patient critical; Sepsis: in-patient severe and critical sepsis; SeConv: sepsis convalescence.