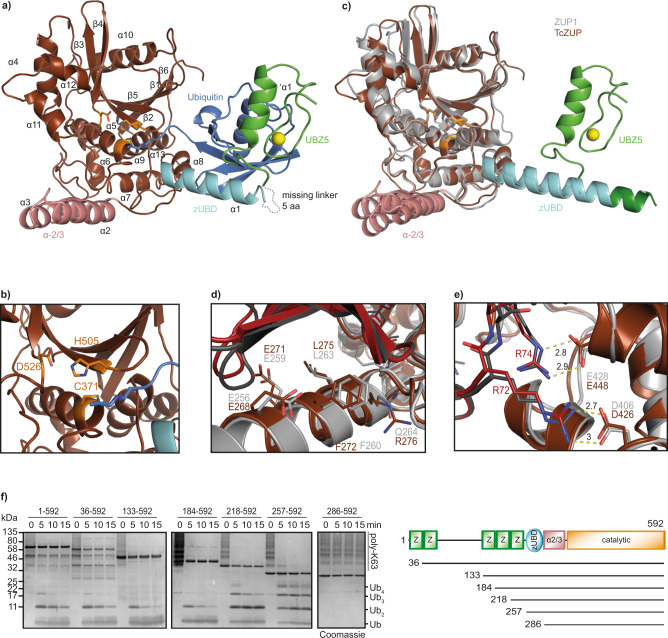
Fig. 3. Crystal structure of T. castaneum ZUP bound to a covalent Ub probe.
a Overview of the crystal structure in cartoon representation. The catalytic core of TcZUP is shown in brown, ubiquitin in blue. The ZnF is colored green, the zUBD region in cyan and the α-2/3 region in red. The catalytic triad is shown as orange sticks. The Zn atom of the ZnF is shown as a yellow sphere. b Magnification of the active site of TcZUP (brown). The catalytic triad is shown as sticks and colored orange. c Structural superposition of TcZUP (brown) and ZUP1 (6EI1, gray). RMS distance is 1.3 Å over 304 residues. The characteristic helical protrusions of ZUP1 and TcZUP are colored cyan (zUBD) and red (α-2/3). The ZnF of TcZUP and the MIU of ZUP1 are colored light and dark green respectively. d Ubiquitin binding by the zUBD is conserved in TcZUP. The zUBD of ZUP1 (light gray) and TcZUP (brown) were superimposed and shown in cartoon representation. Key residues are highlighted as sticks. The corresponding ubiquitins are shown in carton representation and colored dark gray and red, respectively. e Recognition of ubiquitin C terminus by the catalytic core of TcZUP. The TcZUP/ubiquitin (brown/red) and the ZUP1/ubiquitin (light gray/dark gray) complexes are superimposed and shown in cartoon representation. Key residues are highlighted as sticks. Salt bridges are indicated by dotted lines. f Activity of TcZUP FL and truncations lacking the UBDs against K63-linked Ub6+ chains. A mixture of K63-linked poly-ubiquitin chains (Ub6+) are cleaved over time to mono-, di-, tri-, and tetra-ubiquitin. The reaction was stopped after the indicated time points, separated by SDS-PAGE and coomassie stained. Source data are provided as a Source Data file.