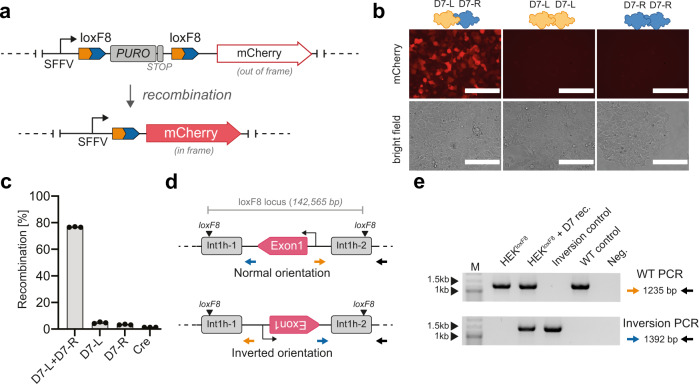
Fig. 3. Activity of the D7 heterodimer in human cells.
a Schematic overview of the integrated reporter construct in HEK293T cells. Expression of mCherry is blocked by the puromycin (PURO) gene. After site-specific recombination the spleen focus-forming virus (SFFV) promoter-driven puromycin resistance cassette is excised, leading to expression of the red-fluorescent mCherry gene. b Fluorescent and brightfield images of transfected HEK293TloxF8 reporter cells with indicated recombinases. Note that mCherry expression is only visible in cells receiving both monomers. 200 µm scale bars are indicated. c Quantification of recombination efficiencies 48 h after transfection of HEK293TloxF8 reporter cells with indicated recombinases, analyzed by flow cytometry (n = 3, biological replicates are shown as dots). Error bars represent standard deviation of the mean (SD). d Schematic overview of a fraction of the F8 gene displaying the PCR primers used to detect the orientation of the loxF8 locus. Exons are displayed in magenta and the repeated regions int1h-1 and int1h-2 are shown in gray. Primer binding sites are indicated with arrows. The position of the loxF8 target sites is indicated. The transcription start site of the F8 gene is depicted by a black arrow. e Gel image of PCR products generated using indicated primer combinations to detect the orientation of the loxF8 locus with and without treatment with the D7 heterodimer. Band sizes of the marker lane (M) are indicated. Inversion control = genomic DNA of patient-derived iPSCs carrying the exon1 inversion. WT control = genomic DNA of HEK293TloxF8 reporter cells (non-treated). Neg. = water used as template for the PCR. Source data are provided as a Source data file.