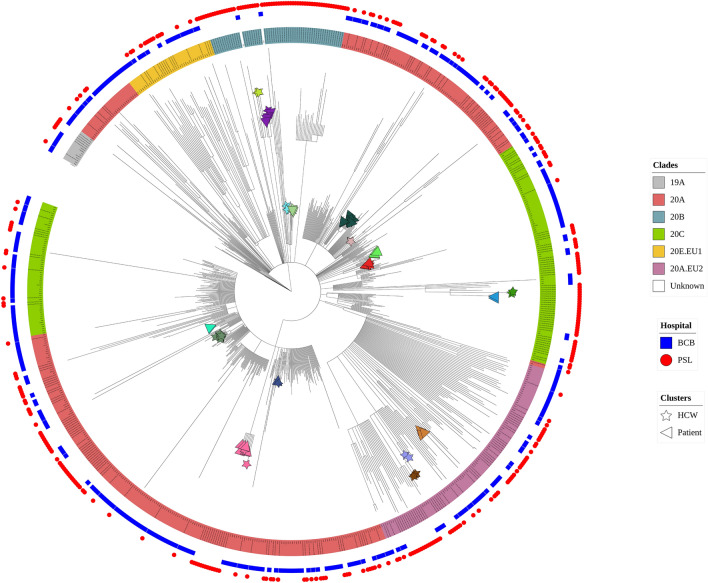
Figure 2.
Phylogenetic tree showing 736 SARS-CoV-2 completes genomes from health care workers and patients from Bichat Claude-Bernard and Pitié-Salpêtrière hospitals, Paris, France, over the year 2020. The maximum likelihood phylogenetic tree was inferred from the alignment of the 736 reconstructed full SARS-CoV-2 genomes and 2932 worldwide sequences extracted from GISAID database, with IQTREE v2.0 with a GTR + G nucleotide substitution model, 1000 ultrafast bootstrap replicates and using Wuhan Hu-1 genome (NC_045512.2) as an outgroup. For greater clarity, 2932 worldwide representative sequences extracted from GISAID database were masked from the phylogenetic tree representation.