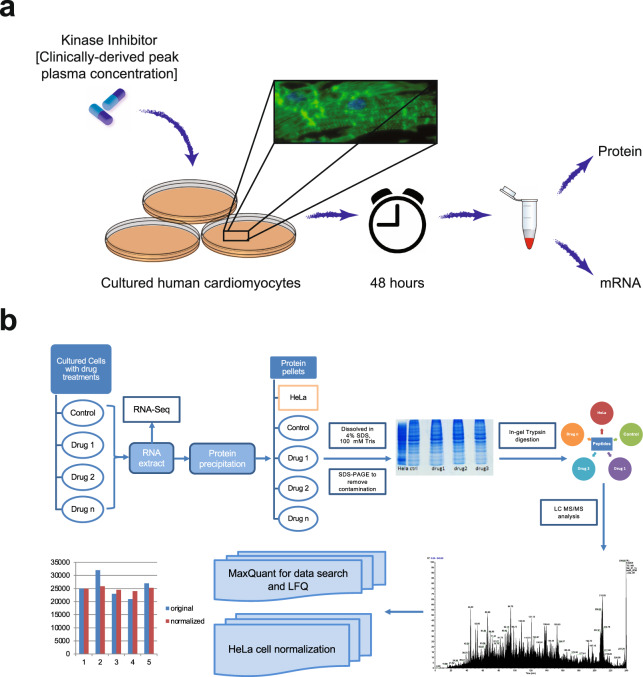
Fig. 1.
LFQ proteomics workflow for the DToxS dataset. (a) Drug and cell treatment design and experimental workflow. (b) After drug treatment, RNA was extracted from cell pellets, and the remaining proteins were recovered via protein precipitation. The protein amount for each sample was carefully estimated from its SDS-PAGE staining intensity as measured relative to a HeLa cell lysate standard. Two micrograms of protein from each sample were analyzed by LC-MS/MS, and the resulting proteins were quantified via LFQ approach using MaxQuant.